Abstract
Pseudosasa usawae is an endemic species in Taiwan, and grows at an altitude of 600–1200 m. In this study, we fully characterized the complete chloroplast genome of P. usawae. The complete chloroplast sequence was 139,660 bp, including large single-copy (LSC), small single-copy (SSC), and a pair of invert repeats (IR) region of 83,271, 12,803, and 21,793 bp. Besides, the plastid genome comprised a total of 129 genes, including protein-coding, tRNA, and rRNA genes as 83, 38, and 8 genes. Phylogenetic analysis reveals that P. usawae is closely associated with Phyllostachys genus clade, sister to the lineage of Phyllostachys.
Pseudosasa usawae (Hayata) Makino and Nemoto Citation1931, a temperate woody bamboo, is a valuable edible bamboo for its shoots in the family Poaceae (Bambusoideae). This species grows at altitudes from 600 to 1200 m which is scattered and associated with broadleaved forests thought the island (Lin Citation1978; Lin Citation2000). In the past, P. usawae was often classified as a species in genus of Arundinaria (Hayata Citation1916), Pleioblastus (Ohki Citation1928), or Pseudosasa (Makino and Nemoto Citation1931). For a long time, Botanist focused on morphological and anatomical to identify the species of Bambusoideae (Leandro et al. Citation2020). In the recent years, molecular biological methods have been used to give detailed information about the phylogenetic of the Bambusoideae, such as Ampelocalamus naibuensis (Zhang and Chen Citation2016), Gelidocalamus xunwuensis (Zhang et al. Citation2019), Acidosasa gigantea (Zheng et al. Citation2020), Bambusa stenoaurita (Xia et al. Citation2021), but not P. usawae. To date, the genetic data of bamboos of Taiwan are very scarce. In this study, we obtained and first reported the complete chloroplast genome of P. usawae.
Fresh leaves as sequencing materials were collected from Taipei Botanical Garden, Taiwan forestry research institute in Taiwan (25°01′54.7″N 121°30′33.3″E). The collection followed the guidelines of the specimen collection of Taiwan Forestry Research Institute. The voucher specimen was deposited at the Herbarium of Department of Forestry and Natural Resources, National Chiayi University, Taiwan (Herbarium code: CHIA, Kun-Cheng Chang/[email protected]) under collection number of K. N. Kung 3149. High-throughput sequencing library preparation and sequencing using the Illumina NovaSeq system (2 × 150 bp paired-end) were performed by Tri-I Biotech, Inc. in New Taipei city, Taiwan. The complete chloroplast genome was performed using CLC Genomic workbench 21 (CLC Inc., Aarhus, Demark) with Pseudosasa cantorii (NC036824) chloroplast genome as the reference sequence. The annotation was predicted by using GeSeq in default settings (Tillich et al. Citation2017) and was deposited in GenBank under the accession number of MZ153211.
The complete chloroplast genome of sample P. usawae is 139,660 bp, a mean coverage of 321.4. The chloroplast genome contained a pair of inverted repeats (21,793 bp each) separated by a large single-copy region (LSC) of 83,271 bp and a small single-copy region (SSC) of 12,803 bp. The overall base composition is 30.6% for A, 30.5% for T, 19.3% for C, 19.6% for G. The GC content of the whole genome is 38.9%. The genome encoded 129 genes, consisting of 83 coding genes, 38 tRNA genes and 8 rRNA genes.
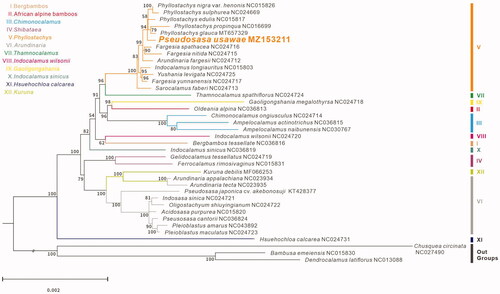
To evaluate the phylogenetic position of P. usawae, additional 37 complete chloroplast genomes in the tribe (). Bambusa emeiensis NC015830, Dendrocalamus latiflorus NC013088 and Chusquea circinata NC024790, the three species in Bambuseae as outgroups, were retrieved from NCBI. All sequences were aligned with MAFFT (Katoh and Standley Citation2013). The maximum-likelihood tree was constructed using 1000 bootstrap replicates by RAxML (Stamatakis Citation2014). The results showed that P. usawae is distantly related to P. cantorii, which is clustered in clade (VI). P. usawae is clustered in the clade (V) with high-support value, as a sister group with the lineage of Phyllostachys genus. The internodes are short between these lineages in the phylogenetic tree, reflecting the probable recent rapid radiation in clade (V).
Ethical approval
This article does not contain any involving human participants or animals studies. The collection followed the guidelines of the specimen collection of Taiwan Forestry Research Institute.
Author contributions
Kuan-Ning Kung and Tsung-Po Chang were involved in the conception and design. Yu-Chun Hsu and Kuan-Ning Kung analyze the data and draft the paper. Kun-Cheng Chang and Shui-Lin Deng revising it critically for intellectual content.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no.MZ153211. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA738262, SRR14826263, and SAMN19716639 respectively.
References
- Hayata B. 1916. Icones Plantarum Formosanarumosa. Vol. VI. Taihoku: Bureau of Productive Industry, Government of Formosa Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Phylogenet Evol. 30(4):772–780.
- Leandro TD, Scatena V, Clark LG. 2020. Comparative leaf blade anatomy and micromorphology in the systematics and phylogeny of Bambusoideae (Poaceae: Poales). Bot J Linn Soc. 192(1):165–183.
- Lin WC. 1978. Bambusoideae. Flora of Taiwan, vol.5 (Poaceae) Taipei: Epoch Publishing Co. Press.
- Lin WC. 2000. Bambusoideae. Flora of Taiwan. 2nd ed., Vol. 5. Taipei: Editorial committee of the Flora of Taiwan, Department of Botany, National Taiwan University Press.
- Makino T, Nemoto K. 1931. Flora of Japan. 2nd ed. Shunyodo shoten: Nihonbashi Tori Sanchome Press.
- Ohki K. 1928. The botanical magazine. Tokyo Bot Soc. 42:520.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Xia H, Liu X, Wang Y, Li X, Wang J, Jin C. 2021. The complete chloroplast genome sequence of Bambusa stenoaurita (Bambusoideae). Mitochondrial DNA B Resour. 6(8):2184–2185.
- Zhang XZ, Chen SY. 2016. Genome skimming reveals the complete chloroplast genome of Ampelocalamus naibunensis (Poaceae: Bambusoideae: Arundinarieae) with phylogenomic implication. Mitochondrial DNA B Resour. 1(1):635–637.
- Zhang YT, Guo CC, Yang GY, Yu F, Zhang WG. 2019. The complete chloroplast genome sequence of Gelidocalamus xunwuensis (Bambusoideae: Arundinarieae): a shrubby bamboo endemic to China. Mitochondrial DNA B Resour. 4(2):3352–3353.
- Zheng X, Yang M, Ding YL, Lin SY. 2020. The complete chloroplast genome sequence of Acidosasa gigantea (Bambusoideae: Arundinarieae): an ornamental bamboo species endemic to China. Mitochondrial DNA B Resour. 5(1):1119–1121.