Abstract
Phalaenopsis stobartiana Reichenbach f. 1877 is mainly distributed in Yunnan province of China and has a high ornamental and breeding value. Here, we reported the chloroplast genome of P. stobartiana. The length of the chloroplast genome was 145,900 bp, encoding 120 genes. The average GC content was 36.8%. Phylogenetic analysis revealed that P. stobartiana and P. wilsonii are closely related. The chloroplast genome could be used for further phylogenetic research, and provide molecular data for future genetic protection and breeding programs.
Phalaenopsis is a popular ornamental flower, commonly known as moth orchids, their blooms last for months. More than 70 origin species within the Phalaenopsis genus and many available cultivars. Phalaenopsis stobartiana is a miniature epiphytic orchid, growing on trunks of trees in dense forests, distributed at 1300 − 1400 m altitude in the Yunnan province, China (Chen and Jeffrey Citation2009). It presents a high ornamental and breeding value, exhibiting green sepals and a distinctive scent (Christenson Citation2001). Unfortunately, with the ruthless collection and habitat loss, P. stobartiana has been listed as the first class protected wild plant in China (http://www.iplant.cn/rep/prot/Phalaenopsis%20stobartiana). In addition, the classification of Phalaenopsis is still confusing due to limited samples species coverage and genetic resources (Deng et al. Citation2015). Therefore, the cp genome information of P. stobartiana would provide valuable molecular data for genetic protection and phylogenetic studies.
The plant samples of P. stobartiana were obtained from Yingjiang County, Yunnan Province, China (24.7 N, 97.9 E), and they were stored in Fujian Agriculture and Forestry University Herbarium (Voucher specimen: GL-YN2019-17A, Yuzhen Zhou, [email protected]). We used the modified CTAB method (Sahu et al. Citation2012) to extract the genomic DNA of P stobartiana from fresh leaves. The DNA was fragmented using Covaris, and the fragments of DNA between 200 and 400 bp were selected. The extracted DNA then underwent end-repairing, phosphorylation, and A-tailing reactions. Thereafter, the fragments were further amplified and circularized. Single stranded DNA circles were formatted as the final library and sequenced using pair-end 100 bp reads on BGISEQ-500. Approximately 14 GB data were generated from the BGI-500 platform and deposited in the GenBank SRA database (accession number: SRX12193139). The data were assembled using SPAdes 3.13.1 software. The assembled circular chloroplast genome was annotated and corrected using GeSeq (Tillich et al. Citation2017) and Geneious Prime v2020.2.1 software. After the annotation results were verified through GB2sequin (Lehwark and Greiner Citation2019), we submitted the chloroplast genome of P. stobartiana to GenBank (accession number: MW531729).
The P stobartiana chloroplast genome was 145,900 bp in size, with an LSC region (85,295 bp), SSC region (10,881 bp), and two IR regions (24,862 bp each). The average GC content was 36.8%. The cp genome encoded 74 protein-coding genes, 38 tRNAs, and 8 rRNAs.
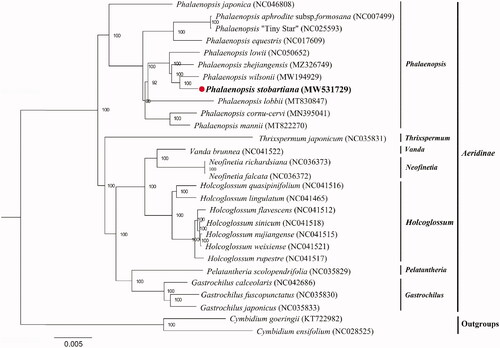
To identify the P. stobartiana’s phylogenetic position, 26 complete chloroplast genomes from Aeridinae and two outgroups were aligned using MAFFT v.7. The maximum likelihood tree was constructed using RAxML-HPC2 on CIPRES with the GTRCAT model and 1000 bootstrap replicates (Stamatakis Citation2014). All the sequences were downloaded from NCBI GenBank. The result indicated that P. stobartiana was sister to P. wilsonii with high bootstrap support ().
Ethical approval
The material involved in the article does not involve ethical conflicts. This study was permitted by the Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization, FAFU, China. All collection and sequencing work was strictly executed under local legislation and related laboratory regulations to protect wild resources.
Author contributions
Conceived and designed the experiments: Y.-Z.Z. Performed the experiments: Y.-H.C., Y. T and Y.-P.Z. Analyzed the data: Y.-H.C., R-Y.Z. and K.Z. Wrote the paper: Y.-H.C. and M.-J.H. Helped to proofread the paper: K.Z., D.-H.P. and Y.-Z.Z. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The plant samples were identified by Dr. Yu-Zhen Zhou, and deposited at the Fujian Agriculture and Forestry University Fuzhou, Fujian, China (Voucher specimen: GL-YN2019-17A).
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW531729.
The associated Bio-Sample, BioProject and SRA numbers are SAMN21439087, PRJNA763377, and SRX12193139, respectively.
Additional information
Funding
References
- Chen SC, Jeffrey JW. 2009. Flora of China. Vol. 25. Beijing: Sciences Press; St. Louis: Missouri Botanical Garden Press; p. 478–483.
- Christenson EA. 2001. Phalaenopsis: a monograph. OR, USA: Timber Press; p. 56–58.
- Deng H, Zhang GQ, Liu ZJ, Wang Y. 2015. A new species and a new combination of Phalaenopsis (Orchidaceae: Epidendroideae: Aeridinae): evidence from morphological and DNA analysis. Phytotaxa. 238(3):243–254.
- Lehwark P, Greiner S. 2019. GB2sequin – a file converter preparing custom GenBank files for database submission. Genomics. 111(4):759–761.
- Sahu SK, Thangaraj M, Kathiresan K. 2012. DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol Biol. 2012:205049.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.