Abstract
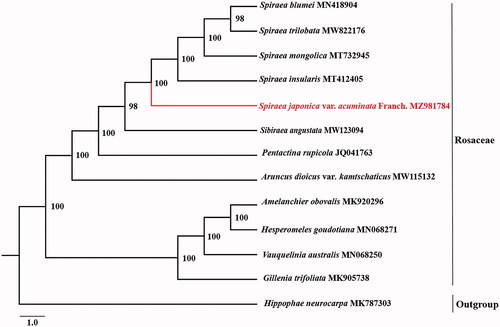
Spirea japonica var. acuminata Franch. (Rosaceae) is a Chinese herbal medicine distributed in southwest and east China. The first complete chloroplast genome of Spirea japonica var. acuminata Franch. was assembled and reported in this study. The genome is 153,822 bp in length and contained 125 encoded genes in total, including 80 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. The phylogenomic analysis showed that Spirea japonica var. acuminata Franch. was closely related to Spirea blumei, Spirea trilobata, Spirea mongolica and Spirea insularis according to the current sampling extent.
Spirea japonica L. f. (1782) is a perennial shrubby species with pink flowers occurring in clusters at the tips of branches. It is widely dissimilar in 14 intraspecific varieties. Spirea japonica var. acuminata Franch. is one variant of Spirea japonica L. f. which is a Chinese herbal medicine classified in the Rosaceae. It is mainly distributed in southwest and east China (He et al. Citation2001; Zhang et al. Citation2006). The extracts from these plants were found to be bioactive for treating cough, anti-inflammation, headache, and analgesia (Li et al. Citation2002). In this study, we assembled and reported the first complete chloroplast genome of Spirea japonica var. acuminata Franch. The cumulative data will provide potential genetic resources for evolution of Rosaceae.
The leaves of Spirea japonica var. acuminata Franch. were collected from Hangzhou, Zhejiang, China (GPS: E120°14′14.95″, N30°23′58.95″). The specimen and extracted DNA was deposited at the College of Life Sciences and Medicine, Zhejiang Sci-Tech University (Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, http://sky.zstu.edu.cn/, Qiu-Ling He and [email protected]) under the voucher number ZSTULSM0001. The total genomic DNA was extracted from its fresh leaves using the CTAB method in accordance with the manufacturer’s instructions. The plastome sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA). In total, ca. 20.6 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. These clean data were de novo assembled to complete chloroplast genome using GetOrganelle (Jin et al. Citation2020).The assembled cp genome was annotated using Geneious v11.1.5 (Drummond Citation2012) with Spirea trilobata plastome (GenBank: MW822176) as a reference.
The full length of Spirea japonica var. acuminata Franch. chloroplast sequence (GenBank Accession No. MZ981784) is 153,822 bp, consisting of a large single copy region (LSC with 82,227 bp), a small single copy region (SSC with 18,907 bp), and two inverted repeat regions (IR with 26,344 bp). The overall GC content of Spirea japonica var. acuminata Franch. chloroplast genome was 36.6% and the GC content of the LSC, SSC, and IR regions are 34.3%, 30.3%, and 42.5%. A total of 125 genes were contained in the genome (80 protein-coding genes, eight rRNA genes, and 37 tRNA genes). Sixteen genes had two copies, which were comprised of fine PCG genes (ndhB, rps7, ycf2, rpl2, rpl23), seven tRNA genes (trnI-CAU, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, trnL-CAA), and all four rRNA species (rrn16, rrn23, rrn4.5, rrn5). In the genome, nine protein-coding genes (atpF, rpl2, ndhB, rps16, rpoC1, clpP, rpl16, petD, petB) had one intron, rps12 and ycf3 gene contained two introns.
To confirm the phylogenetic position of Spirea japonica var. acuminata Franch., we obtained 11 published complete chloroplast genomes of Rosaceae from NCBI. Hippophae neurocarpa was used as outgroup for constructing the phylogenetic tree. The 13 complete cp sequences were aligned using MAFFT v7.3 (Katoh and Standley Citation2013). The maximum-likelihood (ML) phylogenetic analyses were constructed using IQTREE v1.6.7 (Nguyen et al. Citation2015) with 5000 bootstraps under the TVM + F+R3 substitution model. The phylogenetic tree revealed that Spirea japonica var. acuminata Franch. was closely related to Spirea blumei, Spirea trilobata, Spirea mongolica and Spirea insularis according to the current sampling extent ().
Author contributions statement
Qi Wang and Min-min Chen: conception and design, analysis and interpretation of the data, the drafting of the paper. Xia-fang Hu: Data analysis. Rui-hong Wang: supervision. Qiu-ling He: review and editing, funding acquisition. All authors have agreed to be accountable for all aspects of the work..
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper..
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the Accession no. MZ981784. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA759429, SRR15685682, and SAMN21168751, respectively.
Additional information
Funding
References
- Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- He HP, Shen YM, Zhang JX, Zuo GY, Hao XJ. 2001. New diterpene alkaloids from the roots of Spiraea japonica. J Nat Prod. 64(3):379–380.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li L, Shen YM, Yang XS, Zuo GY, Shen ZQ, Chen ZH, Hao XJ. 2002. Antiplatelet aggregation activity of diterpene alkaloids from Spiraea japonica. Eur J Pharmacol. 449(1-2):23–28.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Zhang Z, Fan L, Yang J, Hao X, Gu Z. 2006. Alkaloid polymorphism and ITS sequence variation in the Spiraea japonica complex (Rosaceae) in China: traces of the biological effects of the Himalaya-Tibet Plateau uplift. Am J Bot. 93(5):762–769.