Abstract
The complete mitochondrial genome of the Cephalopholis urodeta (Serranidae: Epinephelinae) was sequenced by next-generation sequencing method on the basis of one female specimen collected from coral reef areas at the Mischief Reef, South China Sea. The mitogenome is 16,593 bp in length, including 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a D-loop region. The overall content of A + T is 55.66%, which is significantly higher than the C + G content (44.34%). Among four bases, C shows the lowest frequency (15.99%). Phylogenetic tree based on the 13 protein-coding genes was constructed for analyzing the position of C. urodeta. The results provide useful insights into further studies on population genetics and phylogenetics of groupers.
As a significant predatory marine fish species, the darkfin hind mainly lives in coral and rocky reefs of the tropical and subtropical oceans worldwide (Heemstra and Randall Citation1993). At the Mischief Reef, South China Sea, Cephalopholis urodeta (Forster, 1801) is one of the dominant fish species on the reef flats and slopes, coexisting with many other groupers, such as Epinephelus chlorostigma (Valenciennes, 1828), E. merra Bloch, 1793, and Lutjanus kasmira (Forsskål, 1775). Though C. urodeta is small in body size, it shows important ecological functions in coral-reef ecosystem (Pinault et al. Citation2014). Several previous studies have been conducted on the stock, taxonomy, evolution, and living habit of C. urodeta (Donaldson 2001; Liu and Sadovy Citation2005; Craig and Hastings 2007; Payet et al. Citation2016). However, limit effort has been made toward the genetic studies of this species so far. Thus, the present study sequenced the mitochondrial genome of C. urodeta, aiming to provide useful molecular markers for further species identification and phylogenetic research.
Samples of C. urodeta were collected from the coral reef areas at the Mischief Reef, South China Sea (9°55′N, 115°32′E) in June 2018. A specimen was deposited at the Key Laboratory of South China Sea Fishery Resources Exploration and Utilization, Ministry of Agriculture and Rural Affairs (Changping Yang, [email protected]) under the voucher number CUF20180601. Total genomic DNA was extracted from the ethanol-preserved muscle tissue using the Marine Animals Genomic DNA Extraction Kit (TIANamp, China). The genomic library construction and next generation sequencing were performed as previously described by Loh et al. (Citation2015). A TruseqTM RNA sample Prep Kit (Illumina Inc., San Diego, CA, USA) was used to construct the whole genome library according to the manufacturer's instructions. The paired-end DNA libraries with insert size of 300–500 bp were PCR-amplified and sequenced on an Illumina HisSeq platform. The clean data without sequencing adapters were then de novo assembled by NOVOplasty software. The assembled genes were annotated using the Blast function of NCBI. The locations of the protein-coding genes (PCGs) and the tRNA genes were determined using ORF Finder via NCBI and the MITOS WebServer, respectively (Wang et al. Citation2018).
The complete mitogenome of C. urodeta is 16,593 bp in length (GenBank accession no. MZ411547), containing 37 mitochondrial genes (13 PCGs, 22 tRNAs, and two rRNAs) and a non-coding region. The gene arrangement order is similar to that of previously reported grouper species C. boenak (Li et al. Citation2014). Twenty-eight of the mitochondrial genes are encoded on the heavy strand, while the remaining nine genes are transcribed from the light strand. The overall A + T content is 55.66%, while the C + G content is 44.34%. Similar to the previously reported grouper species (He et al. Citation2013; Li et al. Citation2014), cytosine shows the lowest frequency (15.99%) among four bases. Three overlapping reading frames are found among the 13 PCGs, 10 bases between atp8 and atp6, 1 base between atp6 and cox3, and 7 bases between nad4L and nad4. The start codon of cox1 and atp6 is GTG and CTG, respectively, while the remaining 11 PCGs use ATG as start codon. Except for cox1, twelve of the 13 PCGs end with TAA terminated codon. The longest gene is cox1 (1551 bp) in all PCGs, whereas the shortest is atp8 (168 bp). Twenty-two tRNAs are determined with 68–73 bp in length. One D-loop region (878 bp) is detected between the trnP and trnF gene. The rrnL and rrnS gene is 957 bp and 1711 bp in length, respectively. They are located between trnF and trnL, and are separated by trnV.
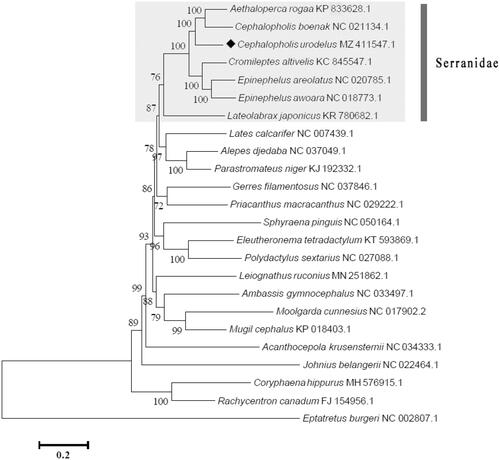
On the basis of 13 PCGs of 24 fish species, a phylogenetic tree (maximum likelihood method, 1000 bootstrap replicates) was constructed to analyze the position of C. urodeta, with the freshwater fish Eptatretus burger (Girard, 1855) as out group. The phylogenetic analysis suggested that C. urodeta was closely related to C. boenak (Bloch, 1790) and Aethaloperca rogaa (Forsskål, 1775), rather than the other three grouper species (Cromileptes altivelis (Valenciennes, 1828), Epinephelus areolatus (Forsskål, 1775), and E. awoara (Temminck and Schlegel, 1842) (). It can be inferred from the phylogenetic tree that the seven species of the family Serranidae formed good monophyly. This complete mitogenome can provide useful genetic information for future studies on phylogeny and the population of the family Serranidae.
Ethical statement
Ethics Committee approval was obtained from the Institutional Ethics Committee of South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences to the commencement of the study.
CRediT authorship contribution statement
Ning Liu: Methodology, Software, Writing – original draft. Dianrong Sun: Formal analysis, Data curation. Binbin Shan: Visualization, Data curation. Yan Liu: Investigation, Writing – review and editing. Liangming Wang: Validation, Data curation. Yingbang Huang: Software, Validation. Changping Yang: Resources, Supervision, Project administration.
Acknowledgments
We thank the editor and anonymous reviewers for their valuable comments on the manuscript.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ411547. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA758836, SRR15667801, and SAMN21037224, respectively.
Additional information
Funding
References
- He BY, Lai TH, Peng ZQ, Wang X, Pan LH. 2013. Complete mitogenome of the Areolate grouper Epinephelus areolatus (Serranidae, Epinephelinae). Mitochondrial DNA. 24(5):498–500.
- Heemstra PC, Randall JE. 1993. FAO Species Catalogue, Vol. 16: Groupers of the World (Family Serranidae, Subfamily Epinephelinae). An annotated and illustrated catalogue of the grouper, rockcod, hind, coral grouper and lyretail species known to date. FAO Fisheries Synopsis 125, FAO, Rome.
- Li JL, Liu M, Wang YY. 2014. Complete mitochondrial genome of the chocolate hind Cephalopholis boenak (Pisces: Perciformes). Mitochondrial DNA. 25(3):167–168.
- Liu M, Sadovy Y. 2005. Habitat association and social structure of the chocolate hind, Cephalopholis boenak (Pisces: Serranidae: Epinephilinae), at Ping Chau Island, northeastern Hong Kong waters. Environ Biol Fish. 74(1):9–18.
- Loh KH, Shao KT, Chen HM, Chen CH, Chong VC, Loo PL, Shen KN, Hsiao CD. 2015. Next generation sequencing yields the complete mitochondrial genome of the Zebra moray, Gymnomuraena zebra (Anguilliformes: Muraenidae). Mitochondrial DNA. 1940–1744:1–2.
- Payet SD, Hobbs JA, DiBattista JD, Newman SJ, Sinclair-Taylor T, Berumen ML, McIlwain JL. 2016. Hybridisation among groupers (genus Cephalopholis) at the eastern Indian Ocean suture zone: taxonomic and evolutionary implications. Coral Reefs. 35(4):1157–1169.
- Pinault M, Bissery C, Gassiole G, Magalon H, Quod JP, Galzin R. 2014. Fish community structure in relation to environmental variation in coastal volcanic habitats. J Exp Mar Biol Ecol. 460:62–71.
- Wang ZF, Wang ZQ, Shi XJ, Wu Q, Tao YT, Guo HY, Ji CY, Bai YZ. 2018. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int J Biol Macromol. 118(Pt A):31–40.