Abstract
In this study, the whole mitochondrial genome of Jacobiasca formosana was sequenced and annotated. The mitogenome is 16,353bp in length, including 37 typical genes and a control region. The 13 length of protein genomes (PCGs) is 10,980bp in size, which encode 3,649 amino acids. All PCGs started with ATN codon and most of PCGs stopped with TAN, except for COX1, COX2 and ND5 used incomplete T as stop codon. The phylogenetic tree results showed that J. formosana belonged to the Empoascini of subfamily Typhlocybinae but different from other species in the subfamily.
The genus of Jacobiasca was established by Dworakowska in 1972 (Dworakowska Citation1972), which included 22 species worldwide and belonged to the tribe Empoascini of subfamily Typhlocybinae (Wang Citation2017). These group of insects are common piercing pests and easy to cause economic losses (Ghauri Citation1963; Oman Citation1969; Theron Citation1974Citation). Jacobiasca formosana was firstly recorded by Paoli in 1932 (Paoli Citation1932) as Empoasca formosana, then was categorized as J. formosana by Dworakowska (Citation1984). Jacobiasca formosana has a wide host range including tea plant (Camellia sinensis), castor oil plant (Ricinus communis), peanut (Arachis hypogaea) and so on. Additionally, the leafhopper could promote the production of Oriental Beauty Tea after it damages the buds and leaves of tea plants (Tsai Citation2014). In this study, sequencing mitochondrial genome of J. formosana will help investigate its taxonomic status and reflect phylogenetic relationships with other species.
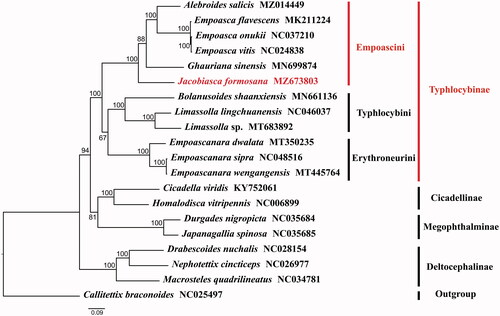
The specimen was collected from Huanglian Mountain, Lvchun County, Yunnan Province, China (N22.67°, E102.33°) by light trapping in August 2018. The leafhopper sampling was permitted by the Institute of Entomology, Guizhou University (The name and number of the project is the National Natural Science Foundation of China [31802002]). Total DNA of J. formosana was extracted from entire body without abdomen using Qiagen DNeasy kit (Qiagen, Venlo, the Netherlands) and following the instruction of manufacture. Male genitalia and voucher specimen’s genome DNA were deposited in the Institute of Entomology of Guizhou University, Guiyang, China (GUGC) with accession number GUGC-IDT-0509064 (Gatherer: Bin Yan, [email protected]). The mitochondrial genome was sequenced by Illumina NovaSeq6000 platform (Berry Genomics, Beijing, China). The reads were assembled using NOVOPlasty 2.7.2 (Dierckxsens et al. Citation2017) and annotated using and Mitoz 2.4-alpha (Meng et al. Citation2019) with the invertebrate mitochondrial genetic codes. The locations of 21 genes were reconfirmed by comparing with the mitogenomes of published Cicadellidae species. The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number MZ673803. Phylogenetic tree was reconstructed using IQ-tree (v1.6.8) with ML analysis based on the amino acids of 13 PCGs of J. formosana and other 20 Cicadellidae species. Callitettix braconoides (Cercopidae) was selected as outgroup.
The circular mitogenome of J. formosana is 16,353 bp in length, including 37 typical genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes), and a control region. The A + T content of genes are 78.8% (A: 38.9%, T: 39.9%, C: 10.8%, G: 10.4%), which is similar to other Typhlocybinae leafhopper mitogenomes (Zhang et al. Citation2020; Shi et al. Citation2020). PCGs encoding 3,649 amino acids, with 10,980bp in size. All PCGs started with ATN codon, and most of them stopped with TAN codon, except for COX1, COX2 and ND5 ended with incomplete T stop codon, respectively. The 22 tRNA are range from 61 bp (trnD, trnA) to 72 bp (trnK), A + T content ranged from 68.6% (trnS) to 87.7% (trnT). The 16 s rRNA and 12 s rRNA are 1,148 and 783 bp in length with an A + T content of 82.1%, respectively. The control region is 2,096 bp in length with an A + T content of 78.4%.
The phylogenetic tree results showed that J. formosana belonged to Empoascini of subfamily Typhlocybinae which is a monophyletic group. Meanwhile, each subfamily was clustered into one clade ().
Author contributions
In this study, Sihan Liu analyzes the data as well as conceives and writes the article; the specimens are collected by Bin Yan; the paper is polished and modified by Rui Shi, Maofa Yang, and Xiaofei Yu, and Yu finally approved the version for publication. All authors agree to be accountable for all aspects of the work.
Acknowledgments
We sincerely thank Dr. Yan Jiang (the Institute of Entomology of Guizhou University) and Dr. Haoxi Li (College of Tobacco Science, Guizhou University, Guiyang, China) for linguistic assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MZ673803. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA773387, SRX12844125, and SAMN22481521, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dworakowska I. 1972. On some oriental and Ethiopian genera of Empoascini (Auchenorrhyncha, Cicadellidae, Typhlocybinae). Bulletin de l'Academie Polonaise des Sciences (Serie Des Sciences Biologiques). 20(1):25–34.
- Dworakowska I. 1984. Studies on Typhlocybinae of Malaysia and Singapore (Homoptera, Auchenorrhyncha, Cicadellidae). Reichenbachia. 22(1):1–21.
- Ghauri MSK. 1963. A new species of Empoasea Walsh (Homoptera: Cicadelloidea) attacking tea in Argetina. Ann Mag Nat Hist. 6(68):465–475.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Oman PW. 1969. New Eupteryginae leafhoppers from Puco Rico (Homoptera Cicadellidae). JAUPR. 21(4):567–572.
- Paoli G. 1932. Specie nuove di Empoasca (Hemiptera-Omoptera) e appunti di corologia. Mem Soc Entomol Italiana. 11:109–122.
- Shi R, Yu XF, Yang MF. 2020. Complete mitochondrial genome of Ghauriana sinensis (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondr DNA Part B. 5(2):1367–1368.
- Theron JG. 1974. A new species of Empoasca (Hemiptera: Cicadellidae), injurious to citrus in South Africa. J Entomol Soc Southern Africa. 37(1):1–3.
- Tsai YF. 2014. Potential of mass rearing and release of Jacobiasca formosana (Hemiptera: Cicadellidae) in Taiwan. PhD thesis. Natl Taiwan Univ. : p5.
- Wang YR. 2017. Molecular phylogeny of the tribe Empoascini (Hemiptera: Cicadellidae: Typhlocybinae). PhD thesis. Northwest A&F University: p11.
- Zhang ZT, Yan SY, Yang WJ, Jin DC. 2020. The complete mitochondrial genome of Gunungidia aurantiifasciata (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondr DNA Part B. 5(2):1393–1394.