Abstract
Tarbinskiellus portentosus (Lichtenstein, 1796) is an agricultural and forestry pest, but people in some places use it as a delicacy. The complete mitogenome of T. portentosus (GenBank accession number MZ427921) is 15, 498 bp in size, including 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs genes, and a noncoding A + T-rich region. The A + T-rich region is located between 12S rRNA and tRNAIle. The base composition of the whole T. portentosus mitogenome is 40.62% for A, 9.87% for G, 32.20% for T, and 17.31% for C, with a high AT content of 72.82%. The phylogeny analysis indicated that T. portentosus had a close relationship with Cardiodactylus muiri. The present data could contribute to further detailed diversity and phylogeographic analysis for this edible cricket.
Tarbinskiellus portentosus (Lichtenstein, 1796), with the common names of short-tail or giant cricket, is an agricultural and forestry pest, but people in some places use it as a delicacy (Soren et al. Citation2021). T. portentosus belongs to Orthoptera: Grylloidea: Gryllidae (Myers et al. Citation2021). The protein content of the adult T. portentosus is 58%, which made it a potential edible insect (Magara et al. Citation2020). T. portentosus belongs to Orthoptera: Gryllidae: Gryllinae, and the reported distribution including China, India, Malaya, Myanmar, Nepal, Pakistan, and Vietnam (Cigliano et al. Citation2020). Elucidating the sequence and structure of T. portentosus mitogenome is important for diversity and phylogeographic analysis of this edible cricket.
The specimen of T. portentosus in this work was obtained from Baise, Guangxi, China (N 23°25′, E 106°38′), and deposited in the insect specimen room (contact person: Cheng-Ye Wang, email: [email protected]) of Research Institute of Resource Insects with voucher number RIRI-w-20200730. Sequencing work of the complete mitogenome of T. portentosus was performed by Illumina Nextseq500 in Beijing Microread Genetics Co., Ltd., with a total data volume 4 G (150 bp Reads). High-quality reads were assembled from scratch using IDBA-UD and SPAdes (Bankevich et al. Citation2012; Peng et al. Citation2012). Protein-coding genes (PCGs) of the T. portentosus mitogenome were identified using BLAST search in NCBI, and tRNA genes were identified using the tRNAscan-SE search server (Schattner et al. Citation2005). The final assembled mitogenome was also verified on the MITOS web server (Bernt et al. Citation2013).
The T. portentosus mitogenome is 15,498 bp in size (GenBank accession number MZ427921), including 13 typical invertebrate PCGs, 22 transfer RNA genes, two ribosomal RNA genes, and a noncoding control region (A + T-rich). The A + T content of the whole T. portentosus mitogenome is 72.82%, showing an obvious AT mutation bias (Nguyen et al. Citation2020). The A + T-rich region exhibits the highest A + T content (77.58%) in the T. portentosus mitogenome.
All the 13 PCGs, 11 PCGs use standard ‘ATN’ as start codons. COX1 use ‘TCG’ as start codon, and ND1 use ‘TTG’ as start codon. As to the stop codon, COX3 and ND5 use ‘T’ as stop codons, and the reminding 11 PCGs have the common mitochondrial stop codon ‘TAA’. Among the 22 tRNAs, 19 tRNAs could be folded into the typical cloverleaf secondary structures; while, tRNASer (GCU) had completely lost the dihydrouridine (DHU) stem; tRNAPhe (GAA) and tRNATyr (GUA) had lost the TφC loop. The 12S rRNA gene is located between the A + T-rich region and tRNAVal, while the 16S rRNA is located between tRNAVal and tRNALeu.
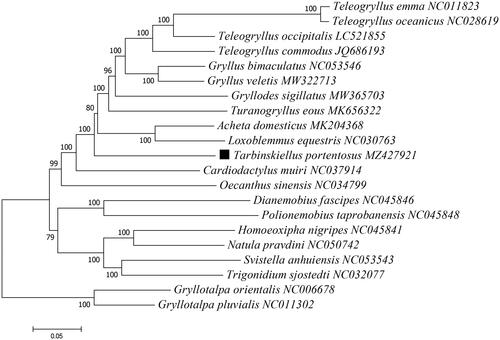
Based on the concatenated PCGs sequences, the maximum-likelihood method was used to construct the phylogenetic relationship between T. Portentosus and 18 other Gryllinae species, with two Gryllotalpinae species used as outgroups (). The phylogenetic analysis was conducted using MEGA version X (Kumar et al. Citation2018). Potential substitution saturation of PCGs data set was assessed using DAMBE5 software (Xia Citation2018), and the outcome (Iss (0.072) < Iss.c (0.784), p < .05) indicated that the substitutions between sequences did not reach saturation and the sequences can be used for subsequent phylogenetic analysis. The phylogeny analysis indicated that T. portentosus had a close relationship with Cardiodactylus muiri (), which add new information to the evolutionary lineage research of T. portentosus (Tantrawatpan et al. Citation2011; He et al. Citation2020; Pradit et al. Citation2021). This mitogenome data might be also valuable for further phylogeography analyses for this edible cricket.
Figure 1. Phylogenetic tree showing the relationship between T. portentosus and 18 other Gryllinae species based on maximum-likelihood method performed using 500 bootstrap replicates. Two Gryllotalpinae species (Gryllotalpa orientalis and G. pluvialis) were used as outgroups. GenBank accession numbers of each sequence were listed in the tree behind their corresponding species names.
Ethical statement
Research procedures in this work did not violate any relevant laws and regulations. No relevant ethics committee or Institution was applicable for the material and data in this study.
Author contributions
Wang CY, Zhao M and Feng Y were involved in the conception and design of the paper. Yang PL, He Z and Sun L were involved in the analysis and interpretation of the data. All authors were involved in the drafting and revising of the paper. All authors agreed the final version to be published and agree to be accountable for all aspects of the work.
Disclosure statement
The authors declare no competing materials in the preparation and execution of this manuscript. The authors are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MZ427921. The associated BioProject, SRA, and bio-sample numbers are PRJNA741082, SRR14902953, and SAMN19844586, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cigliano MM, Braun H, Eades DC, Otte D. 2020. Orthoptera species file. Version 5.0/5.0. http://Orthoptera.SpeciesFile.Org.
- He ZQ, Shen CZ, Wu X, Chen L, Li K. 2020. Report of a new genus Mirigryllus belonging to tribe Modicogryllini, with a new species M. nigrus from Zhejiang, China (Orthoptera: Gryllidae: Gryllinae: Modicogryllini). Zootaxa. 4869(1):zootaxa.4869.1.5.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Magara H, Niassy S, Ayieko MA, Mukundamago M, Egonyu JP, Tanga CM, Kimathi EK, Ongere JO, Fiaboe K, Hugel S, et al. 2020. Edible crickets (Orthoptera) around the world: distribution, nutritional value, and other benefits-A review. Front Nutr. 7:537915.
- Myers P, Espinosa R, Parr CS, Jones T, Hammond GS, Dewey TA. 2021. The animal diversity web. https://animaldiversity.org.
- Nguyen DT, Wu B, Xiao S, Hao W. 2020. Evolution of a record-setting AT-rich genome: indel mutation, recombination, and substitution bias. Genome Biol Evol. 12(12):2344–2354.
- Peng Y, Leung HC, Yiu SM, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Pradit N, Saijuntha W, Pilap W, Suksavate W, Agatsuma T, Jongsomchai K, Kongbuntad W, Tantrawatpan C. 2021. Genetic variation of Tarbinskiellus portentosus (Lichtenstein 1796) (Orthoptera: Gryllidae) in mainland Southeast Asia examined by mitochondrial DNA sequences. Int J Trop Insect Sci.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Soren AD, Choudhury K, Sapruna PJ, Sarma D. 2021. Nutrient and toxic heavy metal assessment of Tarbinskiellus portentosus and Schizodactylus monstrosus consumed by the Bodo tribe in Assam, India. Int J Trop Insect Sci. 41(3):2001–2006.
- Tantrawatpan C, Saijuntha W, Pilab W, Sakdakham K, Pasorn P, Thanonkeo S, Thiha Satrawaha R, Petney T. 2011. Genetic differentiation among populations of Brachytrupes portentosus (Lichtenstein 1796) (Orthoptera: Gryllidae) in Thailand and the Lao PDR: the Mekong River as a biogeographic barrier. Bull Entomol Res. 101(6):687–696.
- Xia X. 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol. 35(6):1550–1552.
