Abstract
Ficus pumila L. is a climbing fig commonly used as an ornamental plant. In this study, we sequenced and assembled the complete chloroplast genome of F. pumila. The complete chloroplast genome of F. pumila is 160,248 bp in length which includes a pair of inverted repeats (IRs) of 25,871 bp separated by a large single-copy (LSC) region of 88,405 bp and a small single-copy (SSC) region of 20,101 bp. The overall guanine–cytosine (GC) content of F. pumila cp genome is 35.98%, while the corresponding values of LSC, SSC, and IR sequences are 33.65, 29.05, and 42.65%, respectively. The phylogenetic tree was shown to be consistent with the traditional morphology-based taxonomy of Moraceae. Five plants from the genus Ficus formed a well-supported monophyletic clade with 100% bootstrap value, and F. pumila is closely related to F. hirta, F. carica, and F. racemosa, with a support value of 97%. The complete chloroplast of F. pumila contributes to the growing number of chloroplast genomes for phylogenetic and evolutionary studies in Moraceae.
Ficus pumila Linnaeus (Citation1753) (Linne Citation1753) is a climbing fig commonly used as an ornamental plant. It is a root climbing evergreen vine attaching to rocks, walls, tree trunks by means of exudations from the aerial roots (Kaur Citation2012). It is reported to be native to East Asia, specifically South China through to Malaysia, but now it is cultivated in numerous countries around the world. The plant is resistant to drought and can also grow under extreme shadow. Therefore, F. pumila has great potential in ecological restoration and vertical greening, and the values of F. pumila in ecological restoration and greening should also attract special attention (Qi et al. Citation2021). Moreover, the plant is believed to have medicinal value. For example, the fruit containing triterpenoids and sterols are used as tonic medicament, antitumor and anti-inflammatory in Chinese folk (Kitajima et al. Citation2000). The leaves containing flavonoid glycosides are traditionally used by some Okinawan elders as beverage or herbal medicines to treat dizziness, diabetes, neuralgia and hypertension. (Liang et al. Citation2012). Therefore, there is an urgent need to obtain genetic and genomic information to advance its systematic research and develop its conservation value of F. pumila.
In this study, the chloroplast genome of F. pumila was sequenced, and its molecular phylogeny and genetic information were determined. The sample of F. pumila was collected from the Guangxi Institute of Botany (GPS: 25°4'46" N, 110°17'52" E) in Guilin, China. The collection of plant materials are in accordance with local regulations and obtain the permission of local authorities. A specimen was deposited at the herbarium of Guangxi Institute of Botany (http://www.gxib.cn/spIBK/, Z. C. Lu, email: [email protected]) under the voucher number IBK00435049. Total genomic DNA was extracted using a modified CTAB (Cetyltrimethyl Ammonium Bromide) method (Doyle and Doyle Citation1987). A total of 6 G raw data from Illumina Hiseq Platform were screened. De novo genome assembly and annotation were conducted by GetOrganelle (Jin et al. Citation2020) and CPGAVAS2 (Shi et al. Citation2019), respectively. The complete cp genome was deposited in GenBank (accession number: MZ351203).
The complete chloroplast genome of F. pumila is 160,248 bp in length which includes a pair of inverted repeats (IRs) of 25,871 bp separated by a large single-copy (LSC) region of 88,405 bp and a small single-copy (SSC) region of 20,101 bp. The overall guanine–cytosine (GC) content of F. pumila cp genome is 35.98%, while the corresponding values of LSC, SSC, and IR sequences are 33.65, 29.05, and 42.65%, respectively.
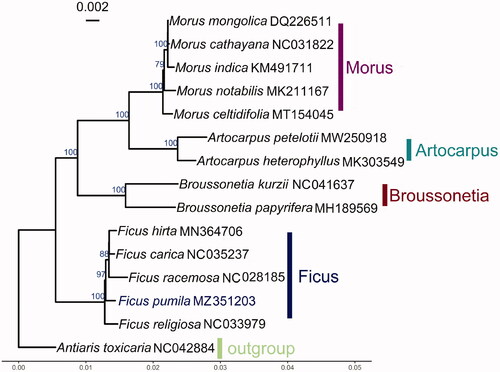
In order to explore the phylogenetic position of F. pumila with other Moraceae, total 14 complete chloroplast genomes published species and one outgroup Antiaris toxicaria (NC042884) were obtained from Genbank. A total of 87 coding sequences were aligned using MUSCLE (Edgar Citation2004). Phylogenetic analysis was conducted using maximum likelihood (ML) method by RAxML version 8.2.12 (Stamatakis Citation2014) with 100 bootstrap replicates. The phylogenetic analysis () showed that five plants from the genus Ficus formed a well-supported monophyletic clade with 100% bootstrap value, and F. pumila is closely related to Ficus hirta, Ficus carica, and Ficus racemosa, with a support value of 97%. The complete chloroplast of F. pumila contributes to the growing number of chloroplast genomes for phylogenetic and evolutionary studies in Moraceae.
Author contributions
JX conceived the original structure of the review. HQ analyzed the data and DY prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ351203. The associated BioProject, SRA, and BioSample numbers are PRJNA763858, SRX12204792, and SAMN21454087 respectively. Treefile of 15 species and genes for phylogenetic analysis were deposited at Figshare: https://doi.org/10.6084/m9.figshare.16628617.v1.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19:11–15.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Depamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):31.
- Kaur J. 2012. Pharmacognostical and preliminary phytochemical studies on the leaf extract of Ficus pumila Linn. J Pharmacog Phytochem. 1:105–111.
- Kitajima J, Kimizuka K, Tanak Y. 2000. Three new sesquiterpenoid glucosides of Ficus pumila fruit. Chem Pharm Bull. 48(1):77–80.
- Liang R-h, Chen J, Liu W, Liu C-m, Yu W, Yuan M, Zhou X-q. 2012. Extraction, characterization and spontaneous gel-forming property of pectin from creeping fig (Ficus pumila Linn.) seeds. Carbohydr Polym. 87(1):76–83.
- Linne C. 1753. Species Plantarum, exhibintes plantas rite cognitas, ad genera relatas, cum differentiis specificis, nominibus tuivialibus, synonymis selectis, locis natalibus, secundum systema sexuale digestas. Verbesina vol, I-II, 902.
- Qi Z-Y, Zhao J-Y, Lin F-J, Zhou W-L, Gan R-Y. 2021. Bioactive compounds, therapeutic activities, and applications of Ficus pumila L. Agronomy. 11(1):89.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.