Abstract
Ligularia stenocephala (Maxim.) Matsum. & Koidz is a widely known edible plant species in Korea. It contains various useful antioxidant compounds and has been developed as a horticultural cultivar blooming showy inflorescence. We report the complete plastid genome (plastome) of Ligularia stenocephala, a collection from Korea. The plastome of L. stenocephala is 151,158 base pairs (bp) long and includes a pair of inverted repeat (IR) regions (24,830 bp each) that are separated by a large single copy (LSC) region (83,265 bp) and a small single copy (SSC) region (18,233 bp). The phylogenetic tree shows that L. stenocephala is closely related to L. fischeri with strong bootstrap support.
The genus Ligularia Cass., belonging to the tribe Senecioneae of Asteraceae, comprises approximately 140 species. It is widely distributed in Europe and Asia, with a primary distribution center in southwestern China (Liu Citation1989; Liu and Illarionova Citation2011). The genus Ligularia is used as a folk medicine to treat influenza, asthma, cough, ulcers, arthritis and tuberculosis in some region of eastern and western Asia (Debnath et al. Citation2017). Therefore, much research has been conducted on the ingredients and efficacy of these medicines. Many phylogenetic studies have shown that Ligularia is a non-monophyletic taxon, demonstrating a common ancestor sharing Cremanthodium and Parasenecio (LCP complex) (Liu et al. Citation2006; Pelser et al. Citation2007; Ren et al. Citation2017). Phylogenomics is a good solution to this phylogenetic conundrum; however, we need more evolutionary evidence of the complex members.
In Korea, Ligularia stenocephala (Maxim.) Matsum. & Koidz.(1910), is commonly known as ‘Gondalbi’. The species is perennial herb and its leaves are consumed as an edible vegetable. It grows on wet, grassy slopes and forest understories, mainly distributed in China, central and southern parts of Japan, the northern part of Taiwan, and several southwestern islands in Korea (Liu Citation1989; Koyama Citation1995). In this study, we report the complete chloroplast (cp) genome sequence of L. stenocephala to provide genomic and genetic sources for further research.
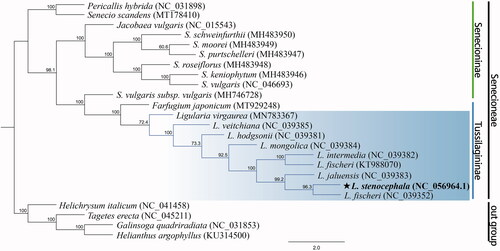
Genomic DNA was extracted from fresh leaves (34°03'42.7"N 125°07'19.9"E; Gageodo, Jeollanam-do Province, Korea) using the DNeasy Plant Mini Kit (Qiagen). The voucher specimen was deposited at the Herbarium of the Kongju National University (KNH, accession number: PMS_180805_001, collected by Minsu Park, E-mail: [email protected]), Kongju, Korea. The DNA library was constructed and sequenced on the MGIseq-2000 platform (LAS, Seoul, Korea) with paired-end reads (150 bp paired-end). The Sequencing generated a total of 88,552,680 raw reads, it was deposited into Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/) with the accession no. SRR13950043. The cp genome sequence of L. stenocephala was constructed by using NOVOPlasty 4.1 (Dierckxsens et al. Citation2017), and for validation the obtained data were mapped with the reference rbcL gene sequence for L. intermedia (Chen et al. Citation2018; MF539930), using Geneious 11.0.4 (Kearse et al. Citation2012). On this genome, 916,240 reads were assembled with a 906.5 × coverage. The annotation was separately performed in Geneious 11.0.4 (Kearse et al. Citation2012), and was manually corrected for the start and stop codons and for the intron/exon boundaries. The annotated cp genome sequence was submitted to GenBank with the accession number of NC_056964.1. A phylogenetic analysis was conducted on the complete chloroplast genome sequences of L. stenocephala and 22 other related species (including 4 outgroups) from the NCBI database and alignments were performed with MAFFT (Katoh and Toh Citation2010). The maximum likelihood (ML) analysis was performed with RAxML v.8.0 (Stamatakis Citation2014) using default parameters and 1,000 bootstrap replicates.
The size of the cp genome of L. stenocephala was 151,158 bp in length, including a large single copy region (LSC) of 83,265 bp, a small single copy region (SSC) of 18,233 bp, and a pair of inverted repeat regions (IRs) of 24,830 bp each. The gene content and order were comparable to other published Ligularia cp genomes (Chen et al. Citation2018). The cp genome contains 132 genes, including 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The overall G + C content of the genome is 37.5, 35.6% in the LSC region, 30.7% in the SSC region, and 43.0% in the IR regions. The phylogenetic tree shows that L. stenocephala is closely related to L. fischeri, with strong bootstrap support (). These data are useful for the phylogenetic and evolutionary studies of LCP complex and Asteraceae.
Ethical approval
The materials used in this study are not included IUCN red list, the collection area is not a protected area. And this article was conducted in compliance with the regulations of the National institute of Biological Resources (NIBR).
Author contributions
Min-Su Park: sample collection, the drafting of the paper.
Won-Bum Cho: data analysis.
Hyun-Do Jang: conception and design of the article.
Chang-gee Jang: literature research and phylogenetic discussion.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. NC_056964.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA713916, SRR13950043, and SAMN18275096 respectively.
Additional information
Funding
References
- Chen X, Zhou J, Cui Y, Wang Y, Duan B, Yao H. 2018. Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol. 9:695–705.
- Debnath T, Kim EK, Nath NCD, Lee KG. 2017. Therapeutic effects of Ligularia stenocephala against inflammatory bowel disease by regulating antioxidant and inflammatory mediators. Food Agric Immunol. 28(6):1142–1154.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26(15):1899–1900.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Koyama H. 1995. Ligularia. In: Iwatsuki K, Yamazaki T, Boufford DE, Ohba H editors. Flora of Japan. Vol. IIIb. Tokyo: Kodansha.
- Liu JQ, Wang YJ, Wang AL, Ohba H, Richard JA. 2006. Radiation and diversification within the Ligularia-Cremanthodium-Parasenecio complex (Asteraceae) triggered by uplift of the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 38(1):31–49.
- Liu SW, Illarionova ID. 2011. Ligularia. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vol. 20–21 (Asteraceae). Beijing and St. Louis: Science Press & Missouri Botanical Garden Press.
- Liu SW. 1989. Ligularia Cass. In Yu B, Zeng GF, editors. Flora Reipublicae Popoularis Sinicae, Vol. 77. Beijing: Science Press.
- Pelser PB, Nordenstam B, Kadereit JW, Watson LE. 2007. An ITS phylogeny of tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon. 56(4):1077–1104.
- Ren C, Hong Y, Wang L, Yang QE. 2017. Generic recircumscription of Parasenecio (Asteraceae: Senecioneae) based on nuclear ribosomal and plastid DNA sequences, with descriptions of two new genera. Bot J Linnean Soc. 184(4):418–443.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.