Abstract
The complete mitogenome of an endemic silkmoth in Taiwan, Antheraea formosana, was determined using Illumina next-generation sequencing. The mitogenome is 15,318 bp in length and consists of 13 protein-coding genes (PCGs), two rRNAs, 22 tRNAs, and one non-coding control region. The overall base composition of the mitogenome showed a high A + T bias, and the A + T content (80.2%) was significantly higher than the G + C content (19.8%). All PCGs use the typical ATN as the initiation codon, with the exception of cox2, which begins with GTG, respectively. The complete mitogenome was used to reconstruct a phylogenetic tree, indicating that A. formosana is more closely related to Antheraea assamensis than other Antheraea species, with 93.19% nucleotide similarity.
Keywords:
The tasar silkmoth (Antheraea Hübner, 1819) belongs to the Lepidoptera family Saturniidae and is of considerable economic importance worldwide (Kitching et al. Citation2018). Antheraea is the largest genus used for silk production and contains more than 35 described species that are widely distributed throughout Asia (Liu et al. Citation2008). Currently, the utilization of silkmoths, including Antheraea pernyi and Antheraea assamensis, for tussah production is greatly prevalent in China, India, and Korea (Peigler Citation1993; Liu et al. Citation2010; Li et al. Citation2017). Antheraea formosana Sonan, 1937, a silk-producing Lepidoptera and an endemic species in Taiwan, is a medium to large-sized silkmoth with a wing span of 110–160 mm, which is distributed in low- and middle-altitude mountainous areas (Chowdhury Citation2004; Wu et al. Citation2020). Because A. formosana behaves as a multivoltine (more than two generations per year), it is a potential alternative candidate for future tussah production in Taiwan. However, there have been no genomic studies on A. formosana. In the present study, we report the whole mitogenome sequence of A. formosana and reconstruct its phylogeny with other Antheraea species.
One A. formosana was collected from Taoyuan District, Kaohsiung (23°09′ N, 120°45′ E) in Taiwan. The collection location in this study is not a privately-owned or protected area, and it is not an endangered or protected species in Taiwan. No permits were required for this study. About an abdominal half of a moth was used to extract total genomic DNA by the use of proteinase-K and phenol-chloroform method (Henry et al. Citation1990). The DNA sample was preserved at the Graduate Institute of Bioresources, National Pingtung University of Science and Technology (Kuo-Hsiang Hung, [email protected]), under the voucher number 2021-AF1. The genomic DNA was used for Illumina library preparation, and subsequently, paired-end reads were sequenced using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA). The raw sequences went through a filtering process to obtain the qualified reads by FASTP v.0.20 (Chen et al. Citation2018), and FLASH v.1.2 was used to merge paired-end reads (Magoč and Salzberg Citation2011). The complete circular mitogenome of A. formosana was assembled de novo using MitoFinder v.1.3 (Allio et al. Citation2020) from a randomly sampled subset of total genomic reads (23,690,470 reads). The assembled mitogenome was annotated using the MITOS2 web server to predict the location of protein-coding regions/genes, tRNAs, and rRNAs (http://mitos2.bioinf.uni-leipzig.de/index.py) (Donath et al. Citation2019). The sequence with annotated features was deposited in GenBank (Accession Number OK078922).
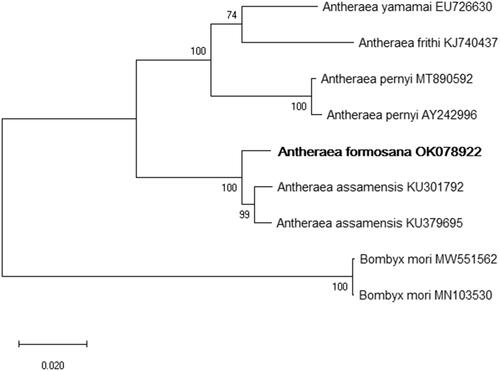
We also inferred phylogenetic relationships based on multiple sequence alignments of the other four Antheraea species mitogenomes, and Bombyx mori was used as an outgroup. The MAFFT online server (Katoh et al. Citation2019) was used to align the mitochondrial sequences, and a maximum-likelihood tree was constructed based on full mitogenome sequences using MEGA X with 1000 bootstrap replicates (Kumar et al. Citation2018).
The mitogenome of A. formosana was 15,318 bp long. It contains 37 genes, comprising 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. The frequencies of adenine, cytosine, guanine, and thymine were 39.3, 12.0, 7.8, and 40.9%, respectively. Therefore, the A + T and G + C contents were 80.2 and 19.8%, respectively. This high A + T bias is similar to that present in other lepidopterans. Twelve PCGs started with a typical ATN codon: four (nad2, cox1, atp8, nad5) with ATT, three (nad3, nad6, cob) with ATA, and five (cox3, atp6, nad4, nad4L, nad1) with ATG. However, cox2 is associated with GTG. This pattern is similar to that of other Antheraea species, except for cox1, in which ATT was the start codon. The cox1 was reported to have CGA as start codons in Antheraea species (Singh et al. Citation2017).
The phylogenetic trees indicated that the Antheraea species were separated into two distinct monophyletic clades. Antheraea formosana is closely related to A. assamensis, with 93.19% nucleotide similarity within the same clade, and other Antheraea species clustered in another clade (). In conclusion, this research provides useful information for further phylogenetic and evolutionary analyses, as well as enriches the mitogenome database of Antheraea species.
Figure 1. The maximum likelihood (ML) phylogenetic tree indicates the relationship between Antheraea formosana and four other Antheraea species. Bombyx mori was used as an outgroup. GenBank accession numbers of each species are listed in the tree. The numbers on the branch lengths are bootstrap values.
Author contributions
An-Ping Cheng, Chi-Chun Huang, Chih-Chiang Wang, I-Ling Lai, and Kuo-Hsiang Hung were involved in the conception and design. An-Ping Cheng and Yu-Tzu Cheng were involved in the collection of materials. Chi-Chun Huang, Yu-Tzu Cheng, Yu-Wei Tseng, Chih-Chiang Wang, and I-Ling Lai were involved in the analysis and interpretation of the data. An-Ping Cheng, Chi-Chun Huang, and Kuo-Hsiang Hung were involved in the drafting of the paper. All authors agreed to be accountable for all aspects of this work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data are available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. OK078922. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA766508, SRR16093533, and SAMN21849811, respectively.
Additional information
Funding
References
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. 2020. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20(4):892–905.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Chowdhury S. 2004. Origin, evolution and distribution of silkworms species. J Assam Sci Soc. 45:43–51.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Henry JM, Raina AK, Ridgway RL. 1990. Isolation of high-molecular-weight DNA from insects. Anal Biochem. 185(1):147–150.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kitching IJ, Rougerie R, Zwick A, Hamilton CA, St Laurent RA, Naumann S, Ballesteros Mejia L, Kawahara AY. 2018. A global checklist of the Bombycoidea (Insecta: Lepidoptera). BDJ. 6(6):e22236.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li W, Zhang Z, Lin L, Terenius O. 2017. Antheraea pernyi (Lepidoptera: Saturniidae) and its importance in sericulture, food consumption, and traditional Chinese medicine. J Econ Entomol. 110(4):1404–1411.
- Liu Y, Li Y, Li X, Qin L. 2010. The origin and dispersal of the domesticated Chinese oak silkworm, Antheraea pernyi, in China: a reconstruction based on ancient texts. J Insect Sci. 10:180.
- Liu Y, Li Y, Pan M, Dai F, Zhu X, Lu C, Xiang Z. 2008. The complete mitochondrial genome of the Chinese oak silkmoth, Antheraea pernyi (Lepidoptera: Saturniidae). Acta Biochim Biophys Sin. 40(8):693–703.
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27(21):2957–2963.
- Peigler RS. 1993. Wild silks of the world. Am Entomol. 39(3):151–162.
- Singh D, Kabiraj D, Sharma P, Chetia H, Mosahari PV, Neog K, Bora U. 2017. The mitochondrial genome of muga silkworm (Antheraea assamensis) and its comparative analysis with other lepidopteran insects. PLOS One. 12(11):e0188077.
- Wu S, Fu CMF, Tzuoo HR, Shih LC, Chang WC, Lin HH. 2020. Formosan entomologist an annotated checklist of macro moths in mid-to high-mountain ranges of Taiwan (Lepidoptera: Macroheterocera). Formosan Entomol. 40:10–83.
