Abstract
The complete mitochondrial genome of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito 2020 (Strain no.: CBS 889.72) was sequenced under the next-generation sequencing platform. It was the second one in the family Mortierellaceae Luerss. 1877. The circular genome was 49,702 bp in size, with a GC content of 20.86%. Gene prediction revealed 15 PCGs, two rRNA genes, 26 tRNA genes, one rnpB gene and seven ORFs. Phylogenetic analyses showed that L. amoeboidea was closely related to Podila verticillate (Linnem.) Vandepol & Bonito 2020.
In the newly proposed framework, the species Mortierella amoeboidea W. Gams 1976 was reclassified into the genus Linnemannia Vandepol & Bonito 2020 as a new combination L. amoeboidea (W. Gams) Vandepol & Bonito 2020 (Vandepol et al. Citation2020). This genus is widely distributed and can be easily isolated from soil, plant debris, insect, etc. (Gams Citation1977). Some species of Linnemannia Vandepol & Bonito 2020 produce polyunsaturated fatty acids, which have potential applications in industrial bioenergy (Zhao et al. Citation2021). Though as many as 120 species are recognized in Mortierellaceae Luerss. 1877, just one has mitogenome available in GenBank (https://www.ncbi.nlm.nih.gov/genome/), which limits the comprehensive and in-depth understanding of this group of fungi. Herein, the mitogenome of L. amoeboidea (W. Gams) Vandepol & Bonito 2020 is analyzed and its phylogenetic position is inferred.
The ex-type strain CBS 889.72 of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito 2020 was collected from Teutoburger Wald, Beller Holz, Germany (51°9′ N, 8°8′ E) and preserved at Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands).
Fungal culture was incubated on PDA for 1 week at 25 °C. Total genomic DNAs were extracted from fresh fungal mycelia using modified CTAB method (Watanabe et al. Citation2010). The duplicate specimen and genomic DNA was deposited at China General Microbiological Culture Collection Center, Beijing, China (http://www.cgmcc.net/, You-Zhi Wang, [email protected]) under the voucher number CBS 889.72. By Illumina HiSeq X-ten sequencing (Nextomics Biosciences, Co., Ltd., Wuhan, China), paired-end libraries with 300 bp inserts were constructed according to the manufacturer’s instructions (Biooscientific, AI™ Paired-End DNA Sequencing Kit). We conducted a quality assessment to obtain clean reads from raw sequencing data by FastQC 0.11.8 (Andrews Citation2010). After that, the mitogenome was assembled from clean data by NOVOPlasty (Dierckxsens et al. Citation2017) with Podila verticillata (Linnem.) Vandepol & Bonito 2020 (NC_006838) as a reference sequence. We annotated the complete mitogenome using the same method as described in previous studies (Zhang et al. Citation2017; Li et al. Citation2021). Briefly, the mitogenome annotation was preliminarily conducted by MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) using the mitochondrial genetic code (genetic code 4) to predict mitogenome organization. The transfer-RNA (tRNA) annotations were identified using tRNAscan-SE v1.3.1 (Lowe and Eddy Citation1997). Intronic and intergenic spacers were searched by ORF Finder (http://www.ncbi.nlm.nih.gov).
The complete mitogenome sequence of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito 2020 was deposited in GenBank under the accession number of MZ411570. It is circular and 49,702 bp in size and have a GC content of 20.86%. The mitogenome contains two ribosomal RNA genes (rnl and rns), 26 tRNA genes, 15 conserved protein-coding genes (atp6, atp8, atp9, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, cox1, cox2, cox3, cob and rsp3), seven free-standing ORFs (orf102, orf188, orf257, orf269, orf277, orf283 and orf412), and one RNA subunit of the mitochondrial RNase P (rnpB). Only one intron, the type of Group IB, was detected in cox1 gene. The result showed that seven PCGs genes (atp6, atp9, cox1, cox2, nad2, nad3 and nad4) are on the forward strand, and other eight genes (atp8, cox3, cob, nad1, nad4L, nad5, nad6 and rps3) are located on the reverse strand.
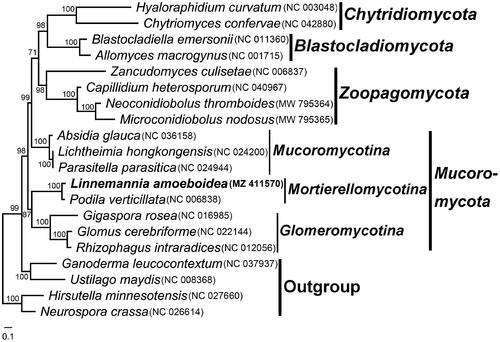
The mitochondrial genome sequences of 19 fungi were downloaded from GenBank for comparison (). Two ascomycetes and two basidiomycetes were chosen as outgroups. Protein sequences from 14 PCGs were used for phylogenetic analyses. Amino acid sequences were aligned with MAFFT v7.051 (Katoh and Standley Citation2013) individually and concatenated with SequenceMatrix v1.7.8 (Vaidya et al. Citation2011). The best model of GTR + I+G for the maximum likelihood (ML) analysis was tested with Modeltest 3.7 (Posada and Crandall Citation1998). The phylogenetic tree was constructed using Maximum Likelihood (ML) method by RAxML 8.1.17 with 1,000 bootstrap replicates (Stamatakis Citation2014). In the clade of Mortierellomycotina Kerst. Hoffm., K. Voigt & P.M. Kirk 2011 (), Linnemannia amoeboidea (W. Gams) Vandepol & Bonito 2020 is most closely related to Podila verticillata (Linnem.) Vandepol & Bonito 2020. Our results also confirm the close relationship of Mortierellomycotina Kerst. Hoffm., K. Voigt & P.M. Kirk 2011 to Glomeromycotina Spatafora & Stajich 2016 and Mucoromycotina Benny 2007 (Spatafora et al. Citation2016; Nie et al. Citation2019), and provide a further understanding of the phylogeny and evolution in basal fungi.
Figure 1. The phylogenetic tree constructed based on 14 mitochondrion encoded proteins. The 14 proteins included oxidase subunits (Cox1, 2, and 3), the apocytochrome b (Cob), ATP synthase subunits (Atp6, Atp8, and Atp9), NADH dehydrogenase subunits (Nad1, 2, 3, 4, 5, 6, and Nad4L). The following other 19 fungal mitogenomes were used in the phylogenetic analysis: Absidia glauca (Ellenberger et al. Citation2016), Allomyces macrogynus (Paquin and Lang Citation1996), Blastocladiella emersonii (Tambor et al. Citation2008), Capillidium heterosporum (Nie et al. Citation2019), Chytriomyces confervae (van de Vossenberg et al. Citation2018), Gigaspora rosea (Nadimi et al. Citation2012), Glomus cerebriforme (Beaudet et al. Citation2013), Hyaloraphidium curvatum (Forget et al. Citation2002), Lichtheimia hongkongensis (Leung et al. Citation2014), Microconidiobolus nodosus (Cai et al. Citation2021), Neoconidiobolus thromboides (Nie et al. Citation2021), Parasitella parasitica (Ellenberger et al. Citation2014), Podila verticillata (Seif et al. Citation2005), Rhizophagus intraradices (Lee and Young Citation2009), and Zancudomyces culisetae (Seif et al. Citation2005). Besides, Ganoderma leucocontextum (NC_037937), Hirsutella minnesotensis (Zhang et al. Citation2016), Neurospora crassa (NC_026614) and Ustilago maydis (NC_008368) were choosen as outgroups. Maximum likelihood bootstrap values (≥70 %) of each clade are indicated along branches. Scale bar indicates substitutions per site. The GenBank accession numbers are behind the Latin names.
Author contributions
BH and XL conceived and designed the experiments. YY analyzed the data and drafted the manuscript. XL improved the manuscript. All authors read and approved the final manuscript. All authors agreed to be accountable for all aspects of the work.
Acknowledgements
We thank Dr. Heng Zhao (Beijing Forestry University) for his assistance with data analyses.
Disclosure statement
The authors have declared that no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ411570. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA741872, SRP325947, and SAMN19911466 respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Babraham Bioinformatics, Babraham Institute.
- Beaudet D, Terrat Y, Halary S, de la Providencia IE, Hijri M. 2013. Mitochondrial genome rearrangements in glomus species triggered by homologous recombination between distinct mtDNA haplotypes. Genome Biol Evol. 5(9):1628–1643.
- Cai Y, Nie Y, Wang ZM, Huang B. 2021. The complete mitochondrial genome of Microconidiobolus nodosus (Entomophthorales: Ancylistaceae). Mitochondrial DNA Part B. 6(6):1743–1744.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Ellenberger S, Burmester A, Wostemeyer J. 2014. Complete mitochondrial DNA sequence of the Mucoralean fusion parasite Parasitella parasitica. Genome Announc. 2(6):e00912–14.
- Ellenberger S, Burmester A, Wostemeyer J. 2016. Complete mitochondrial DNA sequence of the Mucoralean fungus Absidia glauca, a model for studying host-parasite interactions. Genome Announc. 4(2):e00153-16.
- Forget L, Ustinova J, Wang Z, Huss VA, Franz Lang B. 2002. Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol Biol Evol. 19(3):310–319.
- Gams W. 1977. A key to the species of Mortierella. Persoonia. 9(3):381–391.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee J, Young JP. 2009. The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol. 183(1):200–211.
- Leung SY, Huang Y, Lau SK, Woo PC. 2014. Complete mitochondrial genome sequence of Lichtheimia ramosa (syn. Lichtheimia hongkongensis). Genome Announc. 2(4):e00644–14.
- Li Q, Wu P, Li LJ, Feng HY, Tu WY, Bao ZJ, Xiong C, Gui MY, Huang WL. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Nadimi M, Beaudet D, Forget L, Hijri M, Lang BF. 2012. Group I intron-mediated trans-splicing in mitochondria of Gigaspora rosea and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol Biol Evol. 29(9):2199–2210.
- Nie Y, Wang L, Cai Y, Tao W, Zhang YJ, Huang B. 2019. Mitochondrial genome of the entomophthoroid fungus Conidiobolus heterosporus provides insights into evolution of basal fungi. Appl Microbiol Biotechnol. 103(3):1379–1391.
- Nie Y, Wang ZM, Zhao H, Liu XY, Huang B. 2021. Complete mitochondrial genome of Neoconidiobolus thromboides (Entomophthorales: Ancylistaceae). Mitochondrial DNA Part B. 6(7):1840–1841.
- Paquin B, Lang BG. 1996. The mitochondrial DNA of Allomyces macrogynus: the complete genomic sequence from an ancestral fungus. J Mol Evol. 41(5):657–665.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Seif E, Leigh J, Liu Y, Roewer I, Forget L, Lang BF. 2005. Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 33(2):734–744.
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, et al. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia. 108(5):1028–1046.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies . Bioinformatics. 30(9):1312–1313.
- Tambor JH, Ribichich KF, Gomes SL. 2008. The mitochondrial view of Blastocladiella emersonii. Gene. 424(1–2):33–39.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- van de Vossenberg BTLH, Brankovics B, Nguyen HDT, van Gent-Pelzer MPE, Smith D, Dadej K, Przetakiewicz J, Kreuze JF, Boerma M, van Leeuwen GCM, et al. 2018. The linear mitochondrial genome of the quarantine chytrid Synchytrium endobioticum; insights into the evolution and recent history of an obligate biotrophic plant pathogen. BMC Evol Biol. 18(1):136.
- Vandepol N, Liber J, Desirò A, Na H, Kennedy M, Barry K, Grigoriev IV, Miller AN, O'Donnell K, Stajich JE, et al. 2020. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 104(1):267–289.
- Watanabe M, Lee K, Goto K, Kumagai S, Sugita-Konishi Y, Hara-Kudo Y. 2010. Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. J Food Prot. 73(6):1077–1084.
- Zhang YJ, Yang XQ, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101(23–24):8517–8531.
- Zhang YJ, Zhang S, Liu XZ. 2016. The complete mitochondrial genome of the nematode endoparasitic fungus Hirsutella minnesotensis. Mitochondrial DNA Part A. 27(4):2693–2694.
- Zhao H, Lv ML, Liu Z, Zhang MZ, Wang YN, Ju X, Song Z, Ren LY, Jia BS, Qiao M, Liu XY. 2021. High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. Bioenerg Res. 14(4):1196–1206.
