Abstract
The leaf roller, Clepsis pallidana (Fabricius, 1776) (Lepidoptera: Tortricidae), is an important pest of many crops in China. The circular genome is 15,679 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a non-coding AT-rich region. The base composition of the whole mitogenome is 41.2% A, 40.1% T, 11.1% C, and 7.7% G, which shows a strong AT bias. Phylogenetic analysis based on 13 PCGs showed that C. pallidana was closely related to Epiphyas postvittana and clustered within Archipini clade, which was consistent with the traditional classification.
The leaf roller, Clepsis pallidana (Fabricius, 1776), a member of Tortricinae (Lepidoptera: Tortricidae), is a polyphagous moth distributed in the Palearctic Region. Its larvae feed on young leaves and apical buds (Farahbakhsh Citation1961; Liu and Li Citation2002; Sun et al. Citation2021). A total of 34 host plants belonging to 13 families have been recorded for this species from East Asia, Iran, and Europe (Farahbakhsh Citation1961; Liu and Li Citation2002; Sun et al. Citation2021; Yasuda Citation1975; Byun et al. Citation1998; Sohn Citation2002; Jinbo and Nakatani Citation2003; Trematerra and Baldizzone Citation2004). Many of them are important crops, such as Gossypium hirsutum L., Fragaria ananassa Duch., Glycine max (L.), and Malus pumila Mill. To acquire a more comprehensive understanding of Clepsis pallidana, we present the complete mitogenome of C. pallidana as the first mitogenome of the genus Clepsis.
The specimen of this study was neither vertebrates nor regulated invertebrates. And no ethical issues are involved. The study has been granted an exemption from requiring ethical approval by the Committee on the Ethics of Animal Experiments of Northwest University, Xian, China. The specimen used in this study was collected from Taibai County (33°14′N, 107°18′E), Shaanxi Province, China in July 2018 and deposited at Herbarium of Northwest University, China under the voucher number 2018016 (www.nwu.edu.cn; contact Yu Haili, [email protected]). Whole genome sequencing was conducted on Illumina HiSeq 2000 platform with 2 × 150 bp pair-end (GENEWIZ Biotech Co. Ltd., Suzhou, China). After removal of the low-quality reads and adapter region, the trimmed reads were assembled by Novoplasty-NOVOplasty version 3.0 software package (Dierckxsens et al. Citation2017) and annotated by Geneious R8 (Biomatters Ltd, Auckland, New Zealand) under the default parameter settings. PCGs and tRNAs were compared with the sequences in NCBI database to ensure their exact regions. Further, the tRNAscan-SE was used to verify the veracity of tRNA genes (Lowe and Eddy Citation1997).
The complete mitogenome of Clepsis pallidana is 15,679 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a non-coding AT-rich region. The overall base composition of mitogenome is A (41.2%), C (11.1%), G (7.7%), and T (40.1%), with a total of A + T content of 81.3%. The AT-skew and GC-skew of this genome were 0.0156 and −0.1821, respectively. Gene overlaps were found in 5 locations and their total length was 21 bp. The longest overlap was 8 bp in length and resided between trnW and trnC. The length of 22 tRNAs ranged from 64 bp (trnI) to 71 bp (trnK), A + T content ranged from 71.8% (trnK) to 92.6% (trnE). All of these tRNAs can be folded into a canonical cloverleaf secondary structure except for trnS1, and its dihydrouridine (DHU) arm consisted of a simple loop. The 16S rRNA (1,400 bp) was located between trnL1 and trnV, and 12S rRNA (782 bp) resided between trnV and AT-rich region, and their A + T contents were 85.1 and 85.9%, respectively. The non-coding region was located between 12S rRNA and trnM genes with length of 659 bp, and the A + T content was 92.3%. Among the 13 PCGs, nad1, nad4, nad4L, and nad5 were encoded on the light strand (L-strand), while the remaining nine genes were encoded on the heavy strand (H-strand). The A + T content of these 13 PCGs ranged from 72.4% (cox1) to 91.4% (atp8). The cox1 initiated with CGA as the start codon, nad6 started with ATA, atp8 and nad3 started with ATC, nad2 and nad5 started with ATT, and the remaining seven PCGs (cox2, atp6, cox3, nad4, nad4l, cytb, and nad1) started with ATG. Nine PCGs terminated with conventional stop codons TAA, while nad4, nad5, cox1, and cox2 used incomplete codon (T—) as termination codon.
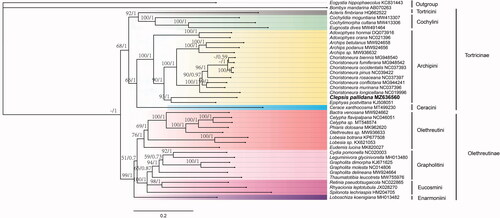
We constructed the phylogenetic relationships based on 13 PCGs from 38 species in Tortricidae, with two outgroup species (). Thirteen PCGs were aligned with the L-INS-i algorithm as implemented in the MAFFT (Katoh and Standley Citation2013). Bayesian inference (BI) was performed with MrBayes version 3.2.6 (Ronquist et al. Citation2012) with the partitioned models determined by PartitionFinder version 2 (Lanfear et al. Citation2017). The maximum-likehood (ML) phylogenetic analysis was performed with 1,000 bootstrap replicates based on the GTR + F+R5 model in the IQ-TREE (Guindon et al. Citation2010; Nguyen et al. Citation2015). As a result, Clepsis pallidana was closely related to Epiphyas postvittana and clustered within Archipini clade. They clustered to the branch of Choristoneura + Archips, and then grouped with two species of Adoxophyes. Monophyly for the clade of Archipini is strongly supported in BI tree, the core of these four genera is unambiguously assigned to the tribe based on a brush of hairs from the venter of the uncus in the male genitalia and a curved, spine-shaped signum with a capitulum in the female genitalia.
Figure 1. Phylogenetic trees inferred from maximum likelihood and Bayesian inference methods based on the 13 PCGs. Numbers separated by a slash (/) on a node represent the bootstrap support and posterior probability, respectively. The specie with mitogenome sequenced in this study is marked in bold. All GenBank accession numbers of each species were listed in the phylogenetic tree. Eogystia hippophaecolus (KC831443) and Bombyx mandarina (AB070263) were used as outgroup.
Author contribution
The authors confirm contribution to the article as follows: study conception and design: Haili Yu, Xueling Song, Ruiqin Dong; data collection: Wenxu Yang; analysis and interpretation of results: Xueling Song, Ruiqin Dong, Wenxu Yang, Haili Yu; draft manuscript preparation: Xueling Song, Haili Yu; revising it critically for intellectual content: Xueling Song. All authors reviewed the results and approved the final version of the manuscript. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genomic sequence data supporting the study's findings are publicly available in NCBI's GenBank https://www.ncbi.nlm.nih.gov/under join number. MZ636560.The associated BioProject, BioSample, and SRA numbers are PRJNA785360, SAMN24563661, and SRR17088790, respectively.
Additional information
Funding
References
- Byun B, Bae Y, Park K. 1998. Illustrated catalogue of Tortricidae of Korea (Lepidoptera). In: Park KT, editor, Insects of Korea [2]. CIS, Korea. p317.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: Assembling organelle genomes from scratch from genome-wide data. Nucleic Acid Res 45(4):e18
- Farabaksh G. 1961. A list of economically important insects and other plants and agricultural enemies of Iran. New Delhi, India: Department of Plant Protection, Ministry of Agriculture; p. 153.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods for estimating maximum likelihood phylogeny: Evaluating the performance of PhyML 3.0 systems biology. 59(3):307–321.
- U.Jinbo, M. Nakatani, 2003. The eastern pale ephemera (Tortricidae, Tortricinae) has a change in wing pattern in Hokkaido. Japanese Pagan 222:427–239.
- Katoh K, Standley DM. 2013. MAFFT Multi-Sequence Comparison Software Release 7: Performance and Usability Improvements. Mol Biol Evol. 30(4):772–780.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: A new approach to selecting partitioned evolutionary models for molecular and morphological phylogenetic analysis. Mol Biol Evol. 34(3):772–773.
- liu yongqiang, li guowei in 2002. insects and animals. lepidoptera: turtle family. volume. 27. beijing: science press; p. 463.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: A procedure for improving the detection of transferred RNA genes in genomic sequences. Nucleic Acid Res 25(5): 955–964
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and efficient stochastic algorithm for estimating maximum likelihood phylogeny. Mol Biol Evol. 32(1):268–274.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu great, Matthew, Justice of the Peace, Welson Baker 2012. MrBayes 3.2: Effective Bayesian phylogenetic inference and model selection in large model spaces. Systems Biology. 61(3):539–542.
- Thorne JC In 2002. A study of the axial phase of the immature stage of the curly-leaved turtle family (Lepidoptera) on the Rosa canina tree in Korea. [Master.] Thesis]. Gangwon-do, South Korea: Gangwon National University; p. 146.
- sun yu, zhang zhibin, cui juan, 2021. astragalus pest species survey plant protection. 47(1):227–232.
- Trematerra P, Baldizzone G. 2004. Records of Totricidae (Lepidoptera) from Krk Island (Croatia). Entomol Croatic. 8:25–44。
- Yasuda T. 1975 Totricinae and Sparganothinae (Lepidoptera: Tortricidae) in Japan (Part II). Boolean University Osaka Branch (B level). 27:79–251.
