Abstract
In the present research, the mitochondrial genome of Ephemera serica was sequenced through next generation sequencing methods and its phylogenetic position in Ephemeroptera was analyzed. Total mitochondrial genome is 15,004 bp in length, and contains 13 protein coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. Mitogenomic phylogeny trees were constructed including 45 species from 13 families. The results show that E. serica is closely related to E. rufomaculata.
Keywords:
The burrowing mayfly genus Ephemera L. (Ephemeroptera: Ephemeridae) contains 68 species worldwide and is mainly distributed in Oriental Region (Hwang and Bae Citation2008). Ephemera species have important position in systematics and phylogeny of Ephemeroptera. However, so far, only four mitochondrial genomes of Ephemera are available to study the phylogenetic relationship of Ephemeroptera (Lee et al. Citation2009; Song et al. Citation2019; Yu et al. Citation2021). Sparse studies conducted on the genus Ephemera have blocked our understanding of the phylogenetic relationship of the genus with other mayfly groups. In this study, we provide a new mitogenome data of Ephemera serica Eaton, 1871, one of most encountered Ephemera species in southern China, for further analyzing the mayfly phylogeny.
Nymphs of Ephemera serica were collected from an inflow stream of the Longdong Reservoir (113.3976°E, 23.2336°N), Guangzhou, China. The voucher specimen of Ephemera serica (the voucher no. E3) was deposited in the Insect Collection, South China Agricultural University (SCAU), Guangzhou, China (Xiaoli Tong, [email protected]). The genomic DNA of the specimen was extracted using phenol–chloroform method. Library prepration was done using TruSeq DNA sample Preparation kit (Vanzyme, China). DNA data were obtained by Illumina Hiseq 2500 (Illumina, USA) with a PE150 strategy (2 × 150 base paired-end reads) and deposited in GenBank (OK018134). Base composition was analyzed in MEGA 7.0 (Kumar et al. Citation2016).
Nesomachilis australica Tillyard, 1924 and Pedetontus silvestrii Mendes, 1993 were selected as outgroups. All 13 PCGs (protein coding genes) and two rRNA genes were aligned individually by MAFFT (Katoh and Standley Citation2013). Gblocks was used to detect the conserved regions with default settings (Talavera and Castresana Citation2007). The best-fit models were selected using PartitionFinder2 (Lanfear et al. Citation2017) by gene types based on Bayesian information criterion (BIC). Phylogenetic analyses were performed using MrBayes 3.2.6 (Ronquist et al. Citation2012) and IQ-TREE (Guindon et al. Citation2010; Minh et al. Citation2013; Nguyen et al. Citation2015). These analyses were all implemented in the PhyloSuite (Zhang et al. Citation2020).
The mitochondrial genome of E. serica was 15,004 bp in length, containing 13 PCGs and two ribosomal RNA genes and 22 transfer RNA genes, but the control region failed to be covered. The overall base contents are 36.2% A, 36.6% T, 16.4% C, and 10.8% G, indicating contents of AT 72.8% and GC 27.2%.
The phylogenetic tree in reveals the relationships of Ephemeroptera that was generated through Bayesian inference (BI) method, and additional maximum likelihood (ML) tree with the identical topology shows only the bootstrap values, except the relationship within Heptageniidae.
Figure 1. Phylogenetic tree of Ephemeroptera based on 13 PCGs and 2 rRNA genes, inferred using MrBayes (BI) and IQ-tree (ML). The values above and below the branches are the Bayesian posterior probability and maximum-likelihood ultrafast bootstrap values, respectively.
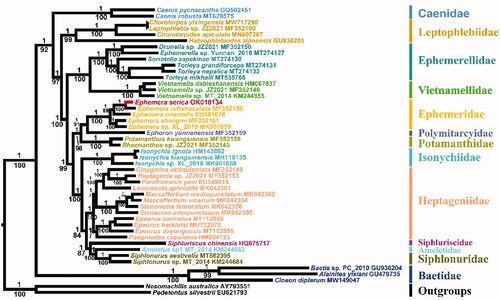
All nodes are strongly supported, except for few nodes in ML analyses with bootstrap values lower than 90. The results show that E. serica is closely related to E. rufomaculata, and three burrowing mayfly families, Ephemeridae, Polymitarcyidae, and Potamanthidae cluster together, which is consistent with the other previous works (Ogden et al. Citation2009; Yu et al. Citation2021). Due to unpublished paper, the data of Siphlonurus immanis (Siphlonuridae) is not included in the current study. Interestingly, S.immanis was clustered a sister clade with Ephemera orientalis (Ephemeridae) in the study of Guan et al. (Citation2021). As a result, Siphlonuridae was divided into two branches, which suggested that Siphlonuridae was recovered as a polyphyly (Guan et al. Citation2021). However, in the present study, Siphlonuridae is supported as monophyletic, which forms a sister clade with Ameletidae. Besides, the unstable position of Isonychiidae still remains (Ogden et al. Citation2009; Guan et al. Citation2021). Resolving these issues require extensive taxon sampling of mitogenomes or larger dataset of related taxa.
Author contributions
Lili Wang and Xiaoli Tong designed the research. Bo Li and Jian Jiang were involved in the sampling and morphological identifing. Lili Wang analyzed the data and wrote the paper. Xiaoli Tong revised the version to be published. All authors agree to be accountable for all aspects of the work.
Ethical approval
Permission for the collection of the sample materials was obtained from South China Insect Diversity Research Center (South China Agricultural University). The approval date was August 1, 2020.
Acknowledgements
Authors are grateful to Jinhong Tian, Xiaofeng Yang, Jianqiang Bi, and Yanping Luo for help with field work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov) under the accession number of OK018134. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA764453, SRR16003088, and SAMN21502591 respectively.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Guan JY, Zhang ZY, Cao YR, Xu XD, Storey B, Yu DN, Zhang JY. 2021. The complete mitochondrial genome of Choroterpes (Euthralus) yixingensis (Ephemeroptera: Leptophlebiidae) and its mitochondrial protein-coding gene expression under imidacloprid stress. Gene. 800:145833.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hwang JM, Bae YJ. 2008. Review of the tropical Southeast Asian Ephemera (Ephemeroptera: Ephemeridae). Aquatic Insects. 30(2):105–126.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Lee EM, Hong MY, Kim MI, Kim MJ, Park HC, Kim YK, Lee IH, Bae CH, Jin BR, Kim I. 2009. The complete mitogenome sequences of the palaeopteran insects Ephemera orientalis (Ephemeroptera: Ephemeridae) and Davidius lunatus (Odonata: Gomphidae). Genome. 52(9):810–817.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ogden TH, Gattolliat JL, Sartori M, Taniczek AH, Soldán T, Whiting MF. 2009. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): combined analysis of morphological and molecular data. Syst Entomol. 34(4):616–634.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Song N, Li X, Yin XM, Li XH, Yin J, Pan PL. 2019. The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships. Sci Rep. 9(1):17765.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Yu D-N, Yu P-P, Zhang L-P, Storey KB, Gao X-Y, Zhang J-Y. 2021. Increasing 28 mitogenomes of Ephemeroptera, Odonata and Plecoptera support the Chiastomyaria hypothesis with three different outgroup combinations. PeerJ. 9(1):e11402.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
