Abstract
Chondrichthyans (sharks, rays and chimeras) are a fascinating and highly vulnerable group of early branching gnathostomes. However, they remain comparatively poorly sampled from the point of view of molecular resources, with deep water taxa being particularly data deficient. The development of long-read sequencing technologies enables the analysis of phylogenetic relationships through a precise and reliable assembly of complete mtDNA genomes. The sequencing and characterization of the complete mitogenome of the opal chimera Chimera opalescens Luchetti, Iglésias et Sellos 2011, using the long-read technique PacBio HiFi is presented. The entire mitogenome was 23,411 bp long and shows the same overall content, i.e. 13 protein-coding genes, 22 transfer RNA and 2 ribosomal RNA genes, as all other examined Chondrichthyan mitogenomes. Phylogenetic reconstructions using all available Chondrichthyan mitogenomes, including 11 Holocephali (chimeras and ratfishes), places C. opalescens within the Chimaeridae family. Furthermore, the results reinforce previous findings, showing the genus Chimera as paraphyletic and thus highlighting the need to expand molecular approaches in this group of cartilaginous fishes.
Chondrichthyans are a monophyletic clade with two sister taxa, the Elasmobranchii (sharks and rays) and Holocephali (chimeras). Their K-selective reproductive traits, such as large body size and slow growth rate (Calis et al. Citation2005; Dagit et al. Citation2007; Kraft et al. Citation2020; Kousteni Citation2021), make them vulnerable to human-mediated threats such as overfishing, particularly elasmobranchs (Cavanagh and Gibson Citation2007; Dulvy et al. Citation2014; Oliver et al. Citation2015; Dulvy and Trebilco Citation2018). Chimaerid are also a frequent by-catch of deep-water fisheries (Blasdale and Newton, Citation1998; Moura et al., Citation2004; Catarino et al. Citation2020). Holocephalans comprise a single surviving order, the Chimaeriformes (Wyffels et al. Citation2014). The described species are allocated into three different families: Callorhinchidae, Rhinochimaeridae and Chimaeridae (Weigmann Citation2016). Furthermore, the family Chimaeridae only includes two genera: Chimera Linnaeus 1758 and Hydrolagus Gill 1862 (Weigmann Citation2016). Recently, several new species have been described (e.g. Iglésias et al. Citation2022), including Chimera opalescens Luchetti et al. Citation2011 from deep-sea assemblages (Luchetti et al. Citation2011). This species is widely distributed in the eastern Atlantic, with records in the British Isles and France (Luchetti et al. Citation2011), on the banks of Greenland, Gorringe and Galicia (Bañon et al. Citation2016; Luchetti et al. Citation2011; Vieira and Cunha Citation2014), Madeira, northwestern slopes of Africa (Freitas et al. Citation2017) and Azores (Catarino et al. Citation2020). The species is listed has Least Concern (LC) according with the Red List of Threatened species of the IUCN (https://www.iucnredlist.org/species/18901743/48862329). However, previous records of C. opalescens were erroneously classified as Chimera monstrosa (Luchetti et al. Citation2011; Catarino et al. Citation2020), due to the similar morphology (Luchetti et al. Citation2011; Didier et al. Citation2012; Freitas et al. Citation2017). This type of problems highlights the critical importance of molecular approaches to support species identification. In this context, the development of long-read sequencing technology has been instrumental, since it allows phylogenetic analysis utilizing complete mtDNA genomes (Satoh et al. Citation2016; Formenti et al. Citation2021).
A female of C. opalescens of 665 mm in total length was captured on 14 October 2020 in the Porcupine Bank (NE Atlantic; Lat:51.1731, Long:-13.5604) at 1037 m depth during the Bottom Trawl Survey PORCUPINE 2020 carried out by the Spanish Institute of Oceanography (IEO, CSIC). Morphological identification was performed onboard, the specimen was frozen, and a muscle tissue sample was stored in absolute ethanol. The specimen is stored at the Spanish Institute of Oceanography in Vigo, with the code voucher C.OPLSCENS_1_P20 (Nair Vilas-Arrondo, [email protected]). The muscle sample is stored at the DNA bank of CIIMAR – Interdisciplinary Center of Marine and Environmental Research with the same voucher code. A small section of the muscle tissue was sent to the Brigham Young University DNA Sequencing Center (BYU), where genomic DNA extraction and whole genome PacBio HiFi library preparation and sequencing were performed, following the manufacturer’s recommendations (Pacific Biosciences; https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-HiFi-SMRTbell-Libraries-using-SMRTbell-Express-Template-Prep-Kit-2.0.pdf). This work has been approved by the CIIMAR ethical committee and by CIIMAR Managing Animal Welfare Body (ORBEA) according to the European Union Directive 2010/63/EU.
The mitochondrial DNA PacBio HiFi (mtDNA PB) reads were filtered by blast search (Altschul et al. Citation1990) against a local built Chondrichthyans mitogenome database and after error corrected using Hifiasm (v.0.13-r308; Cheng et al. Citation2021; Parameters: –write-ec). Subsequently, all reads greater than 20,000 bp were selected and used to perform genome assembly in Unicycler (v.0.4.8.; Parameters: Defaults; Wick et al. Citation2017) a software optimized for circular genome assemblies.
Gene annotation was performed using MITOS2 webserver (Bernt et al. Citation2013) and validated by manual comparison with other chimaerids available at NCBI. For the phylogenetic analysis, all available Chondrichthyan mitogenomes were retrieved from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/, accession date 01/03/2021). Individual alignments for the 13 protein-coding genes (PCG) were produced using MAFFT v7.453 (Katoh and Standley Citation2013) and concatenated using FASconCAT-G (https://github.com/PatrickKueck/FASconCAT-G; final length: 11,431bp). The partition-scheme and the evolutionary best models that fit those schemes and Maximum Likelihood (ML) phylogenetic inference were produced in IQ-TREE (v.1.6.12; Kalyaanamoorthy et al. Citation2017; Nguyen et al. Citation2015). The newly sequenced mitogenome of C. opalescens can be accessed at GenBank (OK638184). The complete mitogenome is 23,411 bp long showing the expected gene composition and arrangement: 13 PCGs, 22 transfer RNA, 2 ribosomal RNA genes, with 14 tRNA, 2 rRNA all PCG (except NAD6) being present in the heavy strand (Satoh et al. Citation2016). We were able to detect and assemble the Holocephali-specific long noncoding insertion present between the tRNAThr and tRNAPro (Inoue et al. Citation2010).
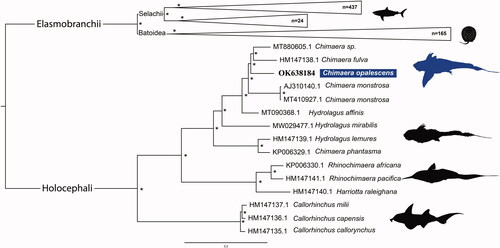
The phylogeny () is divided into two main subclasses: the Holocephali and the Elasmobranchii, reciprocally monophyletic (Boisvert et al. Citation2019).
Within the Holocephali, there are three well-supported clades, Chimaeridae, Rhinochimaeridae and Callorhinchidae (Arnason et al. Citation2001; Inoue et al. Citation2010). As expected, C. opalescens is placed within the family Chimaeridae. However, as previously observed (Gomes-dos-Santos et al. Citation2020, Citation2021), neither Hydrolagus nor Chimera genera were recovered as monophyletic, which highlights the importance of revising the taxonomy. Indeed, previous authors had already suggested that the distinction between Chimera and Hydrolagus based on the presence or absence of a notch separating the anal from the caudal fin, respectively, needed revision (Didier et al. Citation2012 and references therein).
Author contributions
L. F. C. C designed and conceived this work; N. V. -A., F. B., R. B. and E. R. -M. collected the samples; N. V. -A., A. G. S., E. F., L. F. C. C. wrote the first version of the manuscript; NV-A, AGS, MP, AV, DC, AMM, FB, ER-M, R. B., R. R., E. F. and L. F. C. C. carried out the investigation. All authors read, revised and approved the final manuscript.
Disclosure statement
The authors declare no financial interest or benefit from the direct applications of our research.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number OK638184. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA778622, SRR16846874 and SAMN22967859 respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Arnason U, Gullberg A, Janke A. 2001. Molecular phylogenetics of gnathostomous (jawed) fishes: old bones, new cartilage. Zool Scripta. 30(4):249–255.
- Bañon R, Arronte JC, Rodríguez-Cabello C, Piñeiro C-G, Punzón A, Serrano A. 2016. Commented checklist of marine fishes from the Galicia Bank seamount (NW Spain). Zootaxa. 4067(3):293–333.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Blasdale T, Newton AW. 1998. Estimates of discards from two deep water fleets in the Rockall Trough. ICES Document CM 1998/O: 11. Available at: http://www.ices.dk/sites/pub/CM%20Doccuments/1998/O/O1198.pdf.
- Boisvert CA, Johnston P, Trinajstic K, Johanson Z. 2019. Chondrichthyan evolution, diversity, and senses. Springer, Cham; p. 65–91
- Calis E, Jackson EH, Nolan CP, Jeal F. 2005. Preliminary age and growth estimates of the rabbit fish, Chimaera monstrosa, with implications for future resource management. J Northw Atl Fish Sci. 35:15–26.
- Catarino D, Jakobsen K, Jakobsen J, Giacomello E, Menezes GM, Diogo H, Canha Â, Porteiro FM, Melo O, Stefanni S. 2020. First record of the opal chimaera, Chimaera opalescens (Holocephali: Chimaeridae) and revision of the occurrence of the rabbit fish Chimaera monstrosa in the Azores waters. J Fish Biol. 97(3):763–775.
- Cavanagh RD, Gibson C. 2007. Overview of the conservation status of cartilaginous fishes (Chondrichthyans) in the Mediterranean Sea, Gland, Switzerland and Malaga, Spain, IUCN Species Survival Commission Shark Specialist Group.
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18(2):170–175.
- Dagit DD, Hareide N, Clò S. 2007. Chimaera monstrosa. The IUCN Red List of Threatened Species. 2007. e.T63114A12610445.
- Didier DA, Kemper JM, Ebert DA. 2012. Phylogeny, biology, andclassification of extant holocephalans. In: Carrier JC, Musick JA, Heithaus MR, editors. The biology of sharks and their relatives, Vol. 1. Boca Raton, FL: CRC Press; p. 97–122.
- Dulvy NK, Fowler SL, Musick JA, Cavanagh RD, Kyne PM, Harrison LR, Carlson JK, Nk Davidson L, Fordham SV, Francis MP, et al. 2014. Extinction risk and conservation of the world’s sharks and rays. eLife. 3(3):1–34.
- Dulvy NK, Trebilco R. 2018. Size-based insights into the ecosystem role of sharks and rays. In: Carrier J, Heithaus MR, Simpfendorfer CA, eds. Shark research: emerging technologies and applications for the field and laboratory, Vol. 1. Boca Ranton: CRC Press; p. 25–44.
- Formenti G, The Vertebrate Genomes Project Consortium, Rhie A, Balacco J, Haase B, Mountcastle J, Fedrigo O, Brown S, Capodiferro MR, Al-Ajli FO, Ambrosini R, Houde P, Koren S, Oliver K, Smith M, Skelton J, Betteridge E, Dolucan J, Corton C, Bista I, Jarvis ED. 2021. Complete vertebrate mitogenomes reveal widespread repeats and gene duplications. Genome Biol. 22(1):1–22.
- Freitas M, Vieira S, Costa L, Delgado J, Biscoito M, González J. 2017. First records of Chimaera opalescens (Holocephali: Chimaeriformes: Chimaeridae) from Madeira and north-west African coast. Acta Ichthyol Piscat. 47(1):81–84.
- Gomes-dos-Santos A, Arrondo NV, Machado AM, Veríssimo A, Pérez M, Román E, Castro LFC, Froufe E. 2020. The complete mitochondrial genome of the deep-water cartilaginous fish Hydrolagus affinis (deBrito Capello, 1868) (Holocephali: Chimaeridae). Mitochondrial DNA Part B. 5(2):1810–1812.
- Gomes-Dos-Santos A, Vilas-Arrondo N, Machado AM, Veríssimo A, Pérez M, Baldó F, Castro LFC, Froufe E. 2021. Shedding light on the Chimaeridae taxonomy: the complete mitochondrial genome of the cartilaginous fish Hydrolagus mirabilis (Collett, 1904) (Holocephali: Chimaeridae). Mitochondrial DNA B Resour. 6(2):420–422.
- Iglésias SP, Kemper JM, Naylor GJP. 2022. Chimaera compacta, a new species from southern Indian Ocean, and an estimate of phylogenetic relationships within the genus Chimaera (Chondrichthyes: Chimaeridae). Ichthyol Res. 69(1):31–45.
- Inoue JG, Miya M, Lam K, Tay BH, Danks JA, Bell J, Walker TI, Venkatesh B. 2010. Evolutionary Origin and Phylogeny of the Modern Holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 27(11):2576–2586.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kousteni V, Mazzoleni S, Vasileiadou K, Rovatsos M. 2021. Complete mitochondrial DNA genome of nine species of sharks and rays and their phylogenetic placement among modern elasmobranchs. Genes. 12(3):324.
- Kraft DW, Conklin EE, Barba EW, Hutchinson M, Toonen RJ, Forsman ZH, Bowen BW. 2020. Genomics versus mtDNA for resolving stock structure in the silky shark (Carcharhinusfalciformis). PeerJ. 8:e10186.
- Luchetti EA, Iglésias SP, Sellos DY. 2011. Chimaera opalescens n. sp., a new chimaeroid (Chondrichthyes: Holocephali) from the north-eastern Atlantic Ocean. J Fish Biol. 79(2):399–417.
- Moura T, Figueiredo I, Machado PB, Gordo LS. 2004. Growth pattern and reproductive strategy of the holocephalan Chimaera monstrosa along the Portuguese continental shelf. J Mar Biol Ass. 84(4):801–804.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Oliver S, Braccini M, Newman SJ, Harvey ES. 2015. Global patterns in the bycatch of sharks and rays. Marine Policy. 54:86–97.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17(1):719.
- Vieira RP, Cunha MR. 2014. In situ observation of chimaerid speciesin the Gorringe Bank: New distribution records for the north-EastAtlantic Ocean. J Fish Biol. 85(3):927–932.
- Weigmann S. 2016. Annotated checklist of the living sharks, batoids and chimaeras (Chondrichthyes) of the world, with a focus on biogeographical diversity. J Fish Biol. 88(3):837–1037.
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 13(6):e1005595.
- Wyffels J, King BL, Vincent J, Chen C, Wu CH, Polson SW. 2014. SkateBase, an elasmobranch genome project and collection of molecular resources for chondrichthyan fishes. F1000Research. 3:191.