Abstract
Cylicocyclus spp. (Nematoda: Strongylida: Cyathostominae) are the common and important parasitic nematodes found in horses and donkeys worldwide. In this study, the complete mitochondrial genome of Cylicocyclus auriculatus Looss 1900, a representative member of this genus from the donkey in Southwest China was determined using the next-generation DNA sequencing technology. The genome was 13,851 bp in size and consisted of 36 genes including 12 protein-coding genes (atp6, cox1-3, cytb, nad1-6 and nad4L), 22 transfer RNA genes and two ribosomal RNA genes (rrnL and rrnS), as well as two non-coding regions. Phylogenetic analysis showed that C. auriculatus and Cylicocyclus insigne Boulenger 1917 were closely related, and then both grouped with other congeneric species and formed a monophyletic relationship with either species of Cyathostomum, Coronocyclus, Cyathostomum, Cylicostephanus or Cylicodontophorus, demonstrating their phylogenetic stability within Cyathostominae. These cumulative mitochondrial DNA data provide novel and useful genetic markers for molecular diagnostic, systematic and evolutionary biological studies of Cyathostominae nematodes.
Cylicocyclus parasites (Nematoda: Strongylida: Cyathostominae), commonly known as the bursate nematodes of equines, represent a common and severe threat to the livestock breeding industry worldwide (Lichtenfels et al. Citation2008; Zhang Citation2017; Hu et al. Citation2020). There are 13 valid species in the genus Cylicocyclus responsible for such morbidity and socioeconomic burdens. Cylicocyclus auriculatus Looss 1900 is a significant member of the Cylicocyclus nematodes and frequently found in donkeys (Lichtenfels et al. Citation2008; Kuzmina and Kuzmin Citation2008; Bu et al. Citation2009, Citation2013). This parasite inhabits in the large intestine of donkeys and can cause abdominalgia, diarrhea, weight loss and even death. Although there have been substantial advances in morphology and biology of C. auriculatus so far, knowledge gaps to understand this nematode at the molecular level, especially in its genetics and molecular epidemiology, are still not sufficiently explored because of lacking suitable genetic markers (Lichtenfels et al. Citation2008; Bu et al. Citation2013). More importantly, until now the morphology-based classification of Cylicocyclus has been controversial (Lichtenfels et al. Citation2008; Zhang and Kong Citation2002; Gao et al. Citation2021). Under this context, we decoded the mitochondrial genome of C. auriculatus, a representative member of this genus from the Dezhou donkey in Southwest China using the Illumina sequencing technology, as the mitochondrial DNA is not only a rich resource for molecular markers but its complete data can also provide novel insights into phylogenetic relationships of Cylicocyclus (Jex et al. Citation2010; Gao et al. Citation2017; Hu et al. Citation2020; Gao et al. Citation2021).
A total of six parasite specimens were obtained from a Dezhou donkey housed in an original breeding farm at Dazhou (31°92′N, 103°29′E), Sichuan Province of Southwest China, after treatment with pyrantel pamoate. These specimens were identified as adults of C. auriculatus with two males and four females, according to morphological keys of Lichtenfels et al. (Citation2008). One male specimen was used for DNA isolation and the remaining were fixed in 5% formalin solution and archived in the Parasitological Museum of Sichuan Agricultural University (https://dop.sicau.edu.cn/; [email protected] (Yue Xie)) under collection numbers XY2018_28-32. After quality and quantity assessment, ∼2 µg genomic DNA was fragmented to construct a 350-bp paired-end (PE) library, followed by sequencing on an Illumina HiSeq X-TEN platform. The clean reads (∼1.5 Gb) were used to assemble a mitochondrial genome with MITObim (Hahn et al. Citation2013). Gene annotation was achieved using MITOS (Bernt et al. Citation2013). The complete genome sequence was deposited in GenBank under accession number: MZ888509.
The mitochondrial genome of C. auriculatus was 13,851 bp in size and encoded 12 protein-coding genes (atp6, cox1-3, cytb, nad1-6 and nad4L), 22 tRNA genes and two rRNA genes (rrnL and rrnS). All genes were unidirectionally transcribed on the same strand. Among these twelve protein-coding genes, atp6, cytb, cox1-3, nad3, nad5, nad6 and nad4L started with ATT, while nad1, nad2 and nad4 used the TTG as the initiation codon. Correspondingly, except for cox3 and nad2 deduced to end with ‘T’ or ‘TAG’, the remaining genes were predicted to use the TAA as the stop codons. Twenty-two tRNA genes ranged from 54 bp (tRNA(AGN)-Ser) to 63 bp (tRNA-Lys) in size. In addition to tRNA-Ser genes, all had a DHU arm and a TV-replacement loop instead of the TψC arm (Xu et al. Citation2015; Gao et al. Citation2017, Citation2021; Li et al. Citation2019; Qiu et al. Citation2019; Hu et al. Citation2020). Two rRNAs, the small rRNA (rrnS; 700 bp) and large (rrnL; 968 bp) subunits, were located between tRNA-Glu and tRNA(UCN)-Ser and between tRNA-His and nad3, respectively. Two non-coding regions, namely the long non-coding region (LNCR; 273 bp) and short non-coding region (SNCR; 88 bp), were located between tRNA-Ala and tRNA-Pro and between nad4 and cox1, respectively.
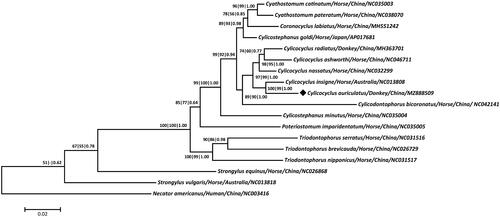
Building on a concatenated amino acid dataset of 12 protein-coding genes from 17 strongyloid parasites, the maximum parsimony (MP), maximum-likelihood (ML) and Bayesian inference (BI) methods were used to construct phylogenetic trees using Necator americanus Stiles 1902 as the outgroup because of its relationship with equine Strongyloidea nematodes (Jex et al., Citation2009; Liu et al., Citation2013; Gao et al., Citation2021). The three identical trees clearly grouped C. auriculatus with Cylicocyclus ashworthi Le Roux 1924, Cylicocyclus insigne Boulenger 1917, Cylicocyclus nassatus Looss 1900 and Cylicocyclus radiates Krecek 2011 and together formed a monophyletic relationship with either species of Cyathostomum, Coronocyclus, Cyathostomum, Cylicostephanus or Cylicodontophorus within Cyathostominae (). Further, C. auriculatus was determined to be more closely related to C. insigne than to other species in Cylicocyclus, with high statistical supports (all bootstrap values ≥99 or =1.00), consistent with recent molecular studies (Hu et al. Citation2020; Gao et al. Citation2021), demonstrating the phylogenetic stability of these bursate nematodes found in equines. Taken together, the sequenced mitochondrial genome of C. auriculatus not only provides novel molecular evidence for its phylogenetic position in Cylicocyclus but also enriches the marker resource for molecular diagnostic, systematic and evolutionary biological studies of strongyloid nematodes.
Figure 1. Phylogeny was inferred from maximum parsimony (MP), maximum-likelihood (ML) and Bayesian inference (BI) analyses based on concatenated amino-acid sequences of 12 mt protein-coding genes of C. auriculatus and other related nematodes. Numbers along the branches represent bootstrap values calculated from different analyses in the order: MP/ML/BI; values < 50% are not shown. The scale indicates an estimate of substitutions per site, using the optimized model setting. The solid black diamond represents the species in this study.
Author contributions
Conceived and designed the research: YX, XZ, XW. Performed the experiments: XL, LW, YC. Analyzed the data and wrote the paper: XL, LW, YX. Revised the paper: YX, XZ, XW. All authors read and approved the final version of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors. The study was reviewed and approved by the Animal Ethics Committee of Sichuan Agricultural University (Sichuan, China; approval no. SYXK 2014-187).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession number MZ888509. The associated BioProject, SRA and Bio-Sample numbers are PRJNA794955, SRR17475793 and SAMN24665689, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bu Y, Niu H, Gasser RB, Beveridge I, Zhang L. 2009. Strongyloid nematodes in the caeca of donkeys in Henan Province. China Acta Parasitol. 54:263–268.
- Bu Y, Niu H, Zhang L. 2013. Phylogenetic analysis of the genus Cylicocyclus (Nematoda: Strongylidae) based on nuclear ribosomal sequence data. Acta Parasitol. 58(2):167–173.
- Gao Y, Qiu JH, Zhang BB, Su X, Fu X, Yue DM, Wang CR. 2017. Complete mitochondrial genome of parasitic nematode Cylicocyclus nassatus and comparative analyses with Cylicocyclus insigne. Exp Parasitol. 172:18–22.
- Gao Y, Wang XX, Ma XX, Zhang ZH, Lan Z, Qiu YY, Wang S, Song MX, Wang CR. 2021. Characterization of the complete mitochondrial genomes of Coronocyclus labiatus and Cylicodontophorus bicoronatus: comparison with Strongylidae species and phylogenetic implication. Vet Parasitol. 290:109359.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hu L, Zhang M, Sun Y, Bu Y. 2020. Characterization and phylogenetic analysis of the first complete mitochondrial genome of Cylicocyclus radiatus. Vet Parasitol. 281:109097.
- Jex AR, Littlewood DT, Gasser RB. 2010. Toward next-generation sequencing of mitochondrial genomes-focus on parasitic worms of animals and biotechnological implications. Biotechnol Adv. 28(1):151–159.
- Jex AR, Waeschenbach A, Hu M, Van Wyk JA, Beveridge I, Littlewood DTJ, Gasser RB. 2009. The mitochondrial genomes of Ancylostoma caninum and Bunostomum phlebotomum-two hookworms of animal health and zoonotic importance. BMC Genomics. 10:79.
- Kuzmina TA, Kuzmin YI. 2008. The community of strongylids (Nematoda, Strongylida) of working donkeys (Equus asinus) in Ukraine. Vest Zool. 2008(42):e18–e23.
- Li Q, Gao Y, Wang XX, Li Y, Gao JF, Wang CR. 2019. The complete mitochondrial genome of Cylicocylus ashworthi (Rhabditida: Cyathostominae). Mitochondrial DNA B Resour. 4(1):1225–1226.
- Lichtenfels JR, Kharchenko VA, Dvojnos GM. 2008. Illustrated identification keys to strongylid parasites (Strongylidae: Nematoda) of horses, zebras and asses (Equidae). Vet Parasitol. 156(1-2):4–161.
- Liu GH, Shao R, Li JY, Zhou DH, Li H, Zhu XQ. 2013. The complete mitochondrial genomes of three parasitic nematodes of birds: a unique gene order and insights into nematode phylogeny. BMC Genomics. 14:414.
- Qiu YY, Zeng MH, Diao PW, Wang XX, Li Q, Li Y, Gao Y, Wang CR. 2019. Comparative analyses of the complete mitochondrial genomes of Cyathostomum pateratum and Cyathostomum catinatum provide new molecular data for the evolution of Cyathostominae nematodes. J Helminthol. 93(5):643–647.
- Xu WW, Qiu JH, Liu GH, Zhang Y, Liu ZX, Duan H, Yue DM, Chang QC, Wang CR, Zhao XC. 2015. The complete mitochondrial genome of Strongylus equinus (Chromadorea: Strongylidae): comparison with other closely related species and phylogenetic analyses. Exp Parasitol. 159:94–99.
- Zhang SY. 2017. Sequence and phylogenetic analysis of COI and ND1 genes for Strongylidae isolated from Equus. Henan Normal University. 1–71.
- Zhang LP, Kong FY. 2002. Review of the systematics of Cyathostominea (Nematoda: strongylidae). Acta Zootax Sin. 27:435–446.
