Abstract
The complete mitochondrial genome of Basiprionota bisignata (Boheman, 1862) (a species of leaf beetles) was successfully sequenced, annotated, and analyzed in this study. This mitochondrial genome is a circular DNA molecule of 16,069 bp in size with 78.5% AT content, including 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and an AT-rich region (control region). The gene order is consistent with the putative ancestral arrangement of insects. All PCGs are initiated by ATN (A/T/C/G) condons and terminated with TAA/G or their incomplete form single T-. All tRNAs can be folded into common clover leaf secondary structures, except for trnS1. The phylogenetic tree was reconstructed using maximum likelihood analysis, and the topology recovered the monophyly of Cassidinae and the sister relationship between Basiprionota and the clade (Thlaspida + Aspidomorph).
The genus (Basiprionota Chevrolat, 1837) belongs to the subfamily Cassidinae of Chrysomelidae, comprising 63 species distributed in Oriental region and border parts of Palearctic and Australopapuan regions (Borowiec Citation1999; Borowiec and Świętojańska Citation2002). In this study, we sequenced the complete mitochondrial genome of Basiprionota bisignata (Boheman, 1862), the first representative of Basiprionota, and performed a phylogenetic analysis among Chrysomelidae with the available mitogenomic sequences.
The samples of B. bisignata (specimen number: GZAF-2021-CC1000) were obtained from Yinjiang County (E108.3666, N28.0846), Guizhou province, China by Huimin Yuan and Ting Wang in May 2021, and stored in Insect Museum of Guizhou Academy of Forestry (URL, Kai Hu and [email protected]), Guiyang. Total genomic DNA was extracted from an adult’s thoracic muscle using the DNeasy Blood and Tissue Kits (Qiagen, Valencia, CA). Based on the high-throughput Illumina Hiseq X platform, total genomic DNA was sequenced. The raw data (3.16 Gb) were assembled using NOVOPlasty version 4.3.1 (Dierckxsens et al. Citation2017) with cox1 sequence from Aspidomorpha difformis (GenBank accession no. MK049862) as the initial seed. The complete mitochondrial genome of B. bisignata was annotated by MITOZ version 1.04 (Meng et al. Citation2019). All 13 protein-coding gene sequences were aligned using MAFFT version 7.394 (Kuraku et al. Citation2013) with L-INSI-I strategy. Maximum likelihood (ML) analysis was conducted using IQ-TREE version 1.6.3 (Nguyen et al. Citation2015) with the optimal model (GTR + I + G for Subset1 (nad3 and atp6), Subset3 (cox1, cytb, cox3, and cox2), and Subset4 (nad1, nad4L, nad4, and nad5); TRN + I + G for Subset2 (nad6 and atp8); TVM + I + G for Subset5 (nad2)) were determined by PartitonFinder2 (Lanfear et al. Citation2017).
The complete mitochondrial genome of B. bisignata is 17,116 bp in length, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and an AT-rich region (control region). The gene order of the newly sequenced mitochondrial genome is consistent with the putative ancestral arrangement of insects (Clary and Wolstenholme Citation1985; Cameron Citation2014). The AT content of the mitochondrial genome is 78.5% (A = 42.8%, T = 35.7%, C = 12.8%, and G = 8.7%), which has a strong AT nucleotide bias. Most PCGs (nad2, cox1, atp8, atp6, nad3, nad4, nad4L, nad6, cytb, and nad1) share typical stop termination TAA/G, whereas cox2, cox3, and nad5 end with incomplete form single T–. Furthermore, all PCGs use ATN (A/T/C/G) as start codon. Length of the 22 tRNAs ranges from 60 bp (trnE) to 68 (trnM). All tRNAs can be folded into common clover-leaf secondary structures, except for trnS1, in which the dihydrourine (DHU) arm formed a simple loop. The size of 16SrRNA and 12SrRNA is 1,259 bp and 740 bp, respectively. The AT-rich region is located between 12SrRNA and trnI, which is 2,515 bp in length with an AT content of 82.8%.
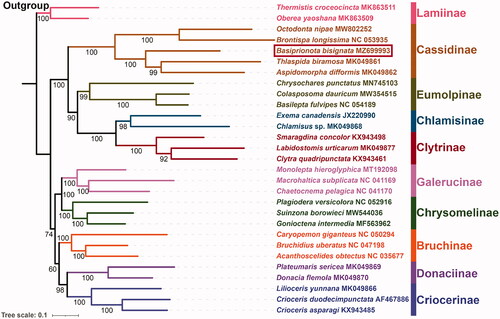
Here, based on the nucleotide data of 13 PCGs from 27 Chrysomelidae species and two outgroup taxa from Cerambycidae, we reconstructed the ML phylogenetic tree (). The phylogenetic relationships among Chrysomelidae are ((Cassidinae + (Eumolpinae + (Chlamisinae + Clytrinae))) + ((Galerucinae + Chrysomelinae) + (Bruchinae + (Donaciinae + Criocerinae)))). In the phylogenetic tree, each subfamily forms a monophyletic cluster with strongly support (BS ≥ 98), consistent with some previous studies (Gómez-Zurita et al. Citation2008; Nie et al. Citation2020). In Cassidinae, the relationships among included genera are inferred as ((Octodonta + Brontispa) + (Basiprionota + (Thlaspida + Aspidomorpha))).
Author contributions
Conceived and designed the experiments: Kai Hu. Performed the experiments: Shaochuan Cheng, Huimin Yuan and Ting Wang. Analyzed the data: Kai Hu. Wrote the paper: Shaochuan Cheng. Helped to proofread the paper: Kai Hu, Huimin Yuan and Ting Wang. All authors have read and agreed to the published version of the manuscript.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of Guizhou Academy of Forestry. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ699993. The associated BioProject, SRAs, and Bio-Sample numbers are PRJNA759086, SRR15686511-SRR15686512, and SAMN21155885, respectively.
Additional information
Funding
References
- Borowiec L, Świętojańska J. 2002. Cassidinae of the world - an interactive manual (Coleoptera: Chrysomelidae). Permanent electronic publication (open in 2002): www.biol.uni.wroc.pl/cassidae/katalog%20internetowy/index.htm.
- Borowiec L. 1999. A world catalogue of the Cassidinae (Coleoptera: Chrysomelidae). Wroclaw (Poland): Biologia Silesiae; p. 1–467.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gómez-Zurita J, Hunt T, Vogler AP. 2008. Multilocus ribosomal RNA phylogeny of the leaf beetles (Chrysomelidae). Cladistics. 24(1):34–50.
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. 2013. Leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41 (Web Server issue):W22–W28.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Nie RE, Andújar C, Gómez‐Rodríguez C, Bai M, Xue HJ, Tang M, Yang CT, Tang P, Yang XK, Vogler AP. 2020. The phylogeny of leaf beetles (Chrysomelidae) inferred from mitochondrial genomes. Syst Entomol. 45(1):188–204.