Abstract
Edenia gomezpompae CRI Eg3 was isolated from the leaves of sweet potato. Its complete mitogenome contains 37,226 bp, with a G + C content of 26.1%. A total of 51 genes were annotated, including 16 protein-coding genes, 33 tRNA genes, and 2 rRNA (s-rRNA, L-rRNA) genes. The most significant character of this mitogenome is its free of group I introns in the CDS regions. Phylogenetic analysis using the mitogenomes of relative fungal species indicated that CRI Eg3 is closely related to Shiraia bambusicola, and they clustered in the Pleosporales lineage. This is the first genome reported in the genus Edenia.
Edenia gomezpompae Gonzalez et al. (Citation2007) was described in 2007 (Gonzalez et al. Citation2007). The type strain was isolated from Callicarpa acuminata leaves, and was identified as an endophytic fungus. Similar strains were later isolated from leaves of Senna alata (Crous et al. Citation2009) and ginseng (Eo et al. Citation2014). The mycelium of Edenia gomezpompae produced naphthoquinone spiroketals with allelochemical, phytotoxic, antiparasitic and in vitro anticancer activities, and have potential application in fungicide (Macias-Rubalcava et al. Citation2008), herbicides (Macias-Rubalcava et al. Citation2014), and medicine (Martinez-Luis et al. Citation2011). Edenia gomezpompae CRI Eg3 was isolated from sweet potato leaves grown in Guangzhou city, Guangdong Province, China with geospatial coordinates N23°23′46″, E 113°26′36″. A specimen was deposited at the Guangdong Microbial Germplasm Preservation Center (http://www.gdmcc.net, Wang Yonghong, [email protected]) under the voucher number GDMCC 3.705. The isolation process includes strict sterilization with 0.1% HgCl2 solution, followed by culturing on potato dextrose agar (PDA) medium. To investigate its taxonomic status and potential biological function, we sequenced its mitochondrial genome.
The genomic DNA was isolated as previously described (Ma et al. Citation2020, Yu et al. Citation2020), and sequenced using Illumina Hiseq 2500 and Nanopore platforms. The subreads were corrected with Canu v1.5 (Koren et al. Citation2017), assembled with wtdbg2 (Ruan and Li Citation2020), and further corrected using pilon (Walker et al. Citation2014). The final circular genome after removing the overlapped sequence was annotated with the MITOS webServer (http://mitos2.bioinf.uni-leipzig.de), and manually corrected using MacVector 13.6. The circular mitochondrial genome has a length of 37,226 bp, with a G + C content of 26.1%. A total of 51 genes were annotated, including 16 protein-coding genes, 33 tRNA genes, 2 rRNA (s-rRNA, L-rRNA) genes. The protein-coding genes include nine for NAD(P)H-quinone oxidoreductases (nad), one for ribosomal protein, one for ATP synthase, three for cytochrome oxidase subunits (coxs), one for cytochrome b (cob), and one for endonuclease. Transfer RNA genes for all 20 amino acids were identified. The most significant character in this mitogenome is that the protein coding sequences are all free of group I introns that are common in fungal mitochondrial genomes (Supplementary Figure S1).
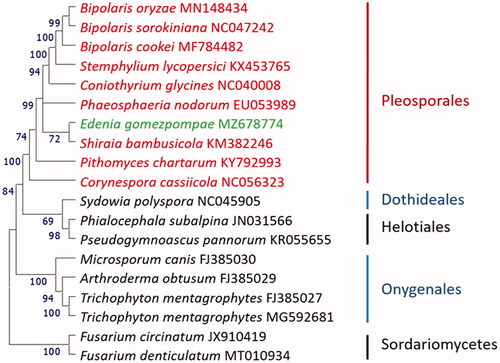
Phylogenetic analysis using 12 peptides encoded by the mitogenomes of relative fungal species indicated that CRI Eg3 is most closely related to Shiraia bambusicola, and they clustered in the Pleosporales lineage ().
Figure 1. Maximum likelihood tree of Edenia gomezpompae and its relative fungal species. Coding DNA sequences shared by all taxa were extracted, translated, and combined in the same order before alignment with Clustal Omega (Sievers and Higgins Citation2018). 12 genes were used in the analysis (nad1, nad2, nad3, nad4, nad4 L, nad5, nad6, atp6, cob, cox1, cox2, cox3). Evolutionary history was inferred by using the maximum likelihood method and JTT matrix-based model (Jones et al. Citation1992), and rooted with two Hypocreales species in Sordariomycetes. Phylogenetic tree was prepared in MEGA11 (Tamura et al. 2016). Bootstrap supports for clades (1000 replicates) are shown above the branches.
Ethical approval
The study involved only a cultivated crop and its endophyte, did not involve any threatened/endangered species. It was exempted from ethical approval and didn’t need any permissions to carry it out.
Author contributions
Lifei Huang isolated the strain and designed the research. Yaojia Mu performed phylogenetic analsyis, Xinxin Zhang cultured and identified the strain. Kaijun Chang and Jiaming Zhang performed genome analysis and wrote the draft. All authors revised the manuscript and approved the final version of the manuscript.
Supplemental Material
Download PDF (152 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ678774. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA761904, SRX12120346, and SAMN21366478, respectively.”
Additional information
Funding
References
- Crous PW, Braun U, Wingfield MJ, Wood AR, Shin HD, Summerell BA, Alfenas AC, Cumagun CJR, Groenewald JZ. 2009. Phylogeny and taxonomy of obscure genera of microfungi. Persoonia. 22:139–161.
- Eo J-K, Choi M-S, Eom A-H. 2014. Diversity of endophytic fungi isolated from Korean ginseng leaves. Mycobiology. 42(2):147–151.
- Gonzalez MC, Anaya AL, Glenn AE, Garcia AS. 2007. A new endophytic ascomycete from El Eden Ecological Reserve, Quintana Roo, Mexico. Mycotaxon. 101:251–260.
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8(3):275–282.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive K-mer weighting and repeat separation. Genome Res. 27(5):722–736.
- Ma S, Yu B, Han B, Tan D, Fu L, Mu Y, Sun X, Zhang J. 2020. The mitochondrion genome of Heveochlorella roystonensis (Trebouxiophyceae) contains large direct repeats. Mitochondrial DNA Part B. 5(2):1355–1356.
- Macias-Rubalcava ML, Hernández-Bautista BE, Jiménez-Estrada M, González MC, Glenn AE, Hanlin RT, Hernández-Ortega S, Saucedo-García A, Muria-González JM, Anaya AL. 2008. Naphthoquinone spiroketal with allelochemical activity from the newly discovered endophytic fungus Edenia gomezpompae. Phytochemistry. 69(5):1185–1196.
- Macias-Rubalcava ML, Ruiz-Velasco Sobrino ME, Meléndez-González C, Hernández-Ortega S. 2014. Naphthoquinone spiroketals and organic extracts from the endophytic fungus Edenia gomezpompae as potential herbicides. J Agric Food Chem. 62(16):3553–3562.
- Martinez-Luis S, Cherigo L, Higginbotham S, Arnold E, Spadafora C, Ibañez A, Gerwick WH, Cubilla-Rios L. 2011. Screening and evaluation of antiparasitic and in vitro anticancer activities of Panamanian endophytic fungi. Int Microbiol. 14(2):95–102.
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17(2):155–158.
- Sievers F, Higgins DG. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27(1):135–145.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS One. 9(11):e112963.
- Yu B, Ma S, Han B, Fu L, Tan D, Sun X, Zhang J. 2020. The complete mitochondrial genome of the rubber tree endophytic alga Heveochlorella hainangensis. Mitochondr DNA Part B. 5(2):1303–1304.
