Abstract
Trigonotis peduncularis (Boraginaceae) is an annual or biannual herb widely distributed in temperate Asia and East Europe. The complete chloroplast genome of T. peduncularis was sequenced by high-throughput technologies and assembled for the first time. The complete chloroplast genome of T. peduncularis was 147,508 bp in length with a GC content of 37.6%, which includes a large single-copy region (80,546 bp), a pair of inverted repeats (24,877 bp), and a small single copy (17,208 bp). GC content of IR regions (43.3%) were higher than LSC (35.5%) and SSC (31.1%) regions. The genome was predicted to encode 130 genes, of which 114 were unique, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. Result from phylogenetic analysis showed that T. peduncularis was sister to Plagiobothrys nothofulvus, and the intergeneric relationships among the six genera sampled in Boraginaceae were well resolved and strongly supported.
The genus Trigonotis Steven (Boraginaceae) consists of approximately 60 species of herbs, mainly distributed in Asia and East Europe (Zhu et al. Citation1995; Weigend et al. Citation2016). There are ca. 40 Trigonotis species recorded in China, and more than 30 are endemic (Zhu et al. Citation1995). Trigonotis peduncularis (Trevis.) Benth. ex Baker & S. Moore 1879 is the most widely distributed species in China, which is also widely distributed in temperate Asia and East Europe. This species has been used as a medical plant for treating stomachache, bleeding, and fracture (Yao et al. Citation2012). Several new compounds have also been discovered in T. peduncularis (Song et al. Citation2008; Otsuka et al. Citation2008).
So far, there is no complete chloroplast genome sequence of Trigonotis available in Genbank. In this study, we reported the complete chloroplast genome sequence of T. peduncularis, which could provide a basis for further study on its genetic diversity, phylogeny, and evolution.
The fresh leaves of T. peduncularis were collected from Nanjing Botanical Garden Mem. Sun Yat-Sen, Nanjing, Jiangsu Province, China (118°49′49′′N, 32°3′32′′E) with the permission from the garden. The voucher specimen (H.M. Li & B. Xue 345) was deposited in the herbaria of the South China Botanical Garden, Chinese Academy of Sciences (IBSC, http://herbarium.scbg.cas.cn/, Feiyan Zeng, [email protected]). Total genomic DNA of T. peduncularis was extracted using the modified CTAB method (Doyle and Doyle Citation1987). Library construction and sequencing were performed by BGI-Shenzhen (Shenzhen, China), using an Illumina HisSeq 2500 Sequencing System following the manufacturer’s instructions. About 3 G data were obtained. The pair-end reads were assembled into a circular contig using NOVOPlasty v.4.2 (Dierckxsens et al. Citation2017). The genome was then annotated using PGA (Plastid Genome Annotator) (Qu et al. Citation2019), and manually corrected in the Geneious v.2020.0.5 (Kearse et al. Citation2012).
The annotated chloroplast genome of T. peduncularis was submitted to GenBank with the accession number MZ911745. The complete chloroplast genome was 147,508 bp in length, which include a large single-copy region, a pair of inverted repeats, and a small single copy, and the sequence lengths were 80,546 bp, 24,877 bp, and 17,208 bp, respectively. The overall GC content is 37.6%, and GC contents of IR regions (43.3%) were higher than that of LSC (35.5%) and SSC (31.1%) regions. The genome was predicted to encode 130 genes, of which 114 were unique, including 80 protein-coding genes, 30 transfer RNA genes, and four ribosomal RNA genes.
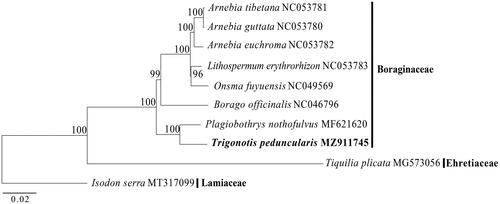
To investigate the phylogenetic position of T. peduncularis, seven chloroplast genome sequences of species from Boraginaceae were downloaded from Genbank for phylogenetic analysis. Chloroplast genome sequences of Tiquilia plicata (Torr.) A.T.Richardson (MG573056) from Ehretiaceae and Isodon serra (Maximowicz) Kudo (MT317099) from Lamiaceae were also downloaded and selected as outgroups. The total ten complete chloroplast genome sequences were aligned using MAFFT v.7.471 (Katoh and Standley Citation2013). A maximum-likelihood analysis was performed by RAxML v. 8.2.12 (Stamatakis Citation2014) under GTR + G model with 1000 bootstrap replicates. Result from phylogenetic analysis showed that T. peduncularis was sister to Plagiobothrys nothofulvus with strong support, and the intergeneric relationships among the six genera sampled in Boraginaceae were also well resolved and strongly supported ().
Authors’ contributions
Jian-hua Wu and Hui-min Li conceived the study and drafted the paper. Jin-man Lei analyzed and interpreted the data. Ze-rui Liang prepared the figure. All authors revised the draft and approved the final draft. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that obtained at this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number of MZ911745. The associated BioProject, Bio-Sample and SRA, numbers are PRJNA767254, SAMN21895048 and SRR16146713, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Otsuka H, Kuwabara H, Hoshiyama H. 2008. Identification of sucrose diesters of aryldihydronaphthalene-type lignan from Trigonotis peducularis and the nature of their fluorescence. J Nat Prod. 71(7):1178–1181.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Song MC, Yang HJ, Kim DK, Baek NI. 2008. A new acyclic diterpene from Trigonotis peduncularis. Bull Korean Chem Soc. 29(11):2267–2269.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Weigend M, Selvi F, Thomas DC, Hilger HH. 2016. Boraginaceae. In: Kadereit JW, Bittrich V, editors. The families and genera of vascular plants 14. Flowering plants. Eudicots. Switzerland: Springer International Publishing; p. 41–102.
- Yao M, Li X, Li WJ, Jia X, Liu XH, Zhao B, Gong J, Mi SF. 2012. Overview of pharmacological research of Trigonotis Stev. J Anhui Agr Sci. 40(9):5130–5131.
- Zhu GL, Riedl H, Kamelin RV. 1995. Boraginaceae. In: Wu ZY, Raven PH, editors, Flora of China, vol. 16. St. Louis (MO): Science Press, Beijing & Missouri Botanical Garden Press; p. 329–427.