Abstract
Anaphalis margaritacea var. yedoensis is a perennial herb adapted to the severe environment of pebbled river banks, where it is frequently found. In this study, we determined the complete chloroplast genome of A. margaritacea var. yedoensis and uncovered its phylogenetic relationships with other members of Gnaphalieae. The total chloroplast genome size of A. margaritaceae var. yedoensis is 153,231 bp, with a large single-copy region (LSC) of 84,981 bp, a small single-copy region (SSC) of 18,481 bp and a pair of inverted repeat (IR) regions of 24,885 bp. A total of 136 genes were annotated, including 39 tRNA genes, 8 rRNA genes, and 89 protein-coding genes. Phylogenetic analysis showed that A. margaritacea var. yedoensis and another Anaphalis species, A. sinica, do not form a monophyletic group, supporting previous phylogenetic studies using some specific regions of cpDNA that showed the genus Anaphalis is non-monophyletic.
Anaphalis margaritacea (L.) Benth. et Hook. f. 1873 (Gnaphalieae, Asteraceae) is a perennial species with a wide distribution ranging from the Himalayas, across East Asia including Japan, to North America (Meng et al. Citation2010). Anaphalis margaritacea var. yedoensis (Franch. et Sav.) Ohwi 1965 is a variety endemic to Japan that has more densely hairy leaves and a better-developed main root than A. margaritacea (Kadota et al. Citation2017). This variety inhabits river banks covered with pebbles exposed to strong sunlight and periodic flooding, and thus the morphological features are thought to be the result of adaptation to the specific environment of its habitat. Genomic information about these tolerance characters may be useful for breeding cultivars belonging to the Asteraceae. Although phylogenetic information about the species is indispensable for such purposes, the taxonomic position of A. margaritacea var. yedoensis has not yet been elucidated. Thus, our goal for this study was to determine the complete chloroplast genome of A. margaritacea var. yedoensis and use it to examine the phylogenetic position of the Gnaphalieae.
We collected an individual of A. margaritacea var. yedoensis from a population (38˚39′06″06 N, 140°52′58″74E) beside the Eai River, Miyagi Prefecture, Japan and cultivated it in the Botanical Gardens, Tohoku University. Collections of the species used in this study are not prohibited by any regulations including our university and regional, national, or international ones. In addition, all the collections were conducted outside legally protected areas. A voucher specimen (Taishi Hoson L8) was deposited in the herbarium of the Botanical Gardens, Tohoku University (TUS; contact Takuro Ito: [email protected]). Fresh leaves were sampled from this cultivated plant and total genomic DNA was extracted using the CTAB method (Doyle and Doyle Citation1987). The purified DNA was sent to a company (Macrogen: Tokyo, Japan) where it was used for paired-end 150 bp sequencing using the Illumina Hiseq X platform. The raw data (83,137,468 reads) were assembled by NOVOplasty (Dierckxsens et al. Citation2017) and annotated by Geseq (Tillich et al. Citation2017). The annotated complete genome sequence of A. margaritacea var. yedoensis was submitted to DDBJ under the accession number LC656264.
The whole cp genome of A. margaritacea var. yedoensis was 153,231 bp in length, with a large single-copy region (LSC) of 84,981 bp, a small single-copy region (SSC) of 18,481 bp and a pair of inverted repeat (IR) regions of 24,885 bp. GC content of the whole cp genome was 37.11%. A total of 136 genes were annotated, including 39 tRNA genes, 8 rRNA genes, and 89 protein-coding genes.
To reveal the phylogenetic position of A. margaritacea var. yedoensis within Gnaphalieae, a phylogenetic analysis was performed with the complete chloroplast genome sequence of A. margaritacea var. yedoensis and other species of Gnaphalieae including the Anaphalis species examined previously (A. sinica Hance 1874). Following previous phylogenetic research using three regions of cpDNA (Fu et al. Citation2016), Artemisia montana (Nakai) Pampan 1965 and Chrysanthemum indicum (L.) des monl. 1888 belonging to Anthemidiae and Astereae, respectively, were used as outgroups (). The sequences were aligned using MAFFT 7 (Katoh and Standley Citation2013). RAxML version 8.2.12 (Stamatakis Citation2014) was used to construct the maximum likelihood tree under the TPM1uf + G4 model that was determined to be the best-fitting model by ModelTest-NG (Darriba et al. Citation2020); bootstrap probability values (BS) were calculated from 1000 replicates.
Figure 1. Phylogenetic position of Anaphalis margaritacea var. yedoensis revealed by a maximum-likelihood tree based on complete chloroplast genomes. Artemisia montana and Chrysanthemum indicum are included as outgroups. The sample used in this study is shown in boldface type. Numbers at nodes indicate bootstrap support values.
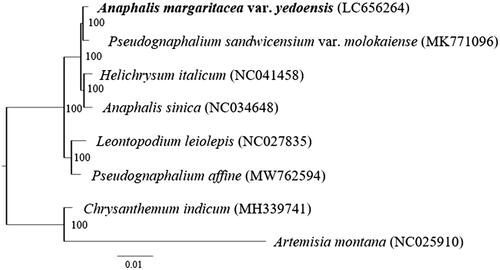
The phylogenetic analysis showed that the two Anaphalis species did not form a monophyletic group while it strongly supported the conclusions that A. margaritacea var. yedoensis and Pseudognaphalium sandwicensium var. molokaiense (O.Deg. & Sherff) W.L.Wagner, 1999 and A. sinica and Helichrysum italicum (Roth) G. Don fil. 1830 were monophyletic with very high bootstrap values (both BS: 100). These results support a previous phylogenetic study showing that there are two phylogenetic groups within Anaphalis (Nie et al. Citation2013) and another study showing that the genera Helichrysum, Anaphalis, and Pseudognaphalium are not monophyletic in a cpDNA tree (Galbany-Casals et al. Citation2014). By contrast, in the latter study, the phylogenetic tree based on nrDNA sequences showed that these genera are monophyletic (Galbany-Casals et al. Citation2014). We also sequenced ITS and ETS regions of A. margaritacea var. yedoensis and reconstructed a phylogenetic tree of the taxa closely related to this variety. As the result, the species was included in a clade comprised from other Anaphalis species (data not shown), supporting the previous results of nrDNA phylogeny (Galbany-Casals et al. Citation2014). The incongruence between the phylogenies based on the cpDNA and nrDNA sequences may have resulted from introgression and/or incomplete lineage sorting. In future studies, it will be necessary to test past gene flow among the genera and to reconsider their taxonomic treatments.
Authors’ contribution
All authors conceptualized and designed this study. TH and TI analyzed and interpreted the data. TH wrote the draft, and MM and TI revised it critically. All authors approved the final version and agreed to be accountable for all aspects of this work.
Acknowledgment
We thank all members of the Botanical Gardens, Tohoku University for their support for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Date availability statement
The data that support the findings of this study are openly available in DDBJ (accession no. LC656264) at http://www.ddbj.nig.ac.jp.The associated BioProject, SRA, and Bio-Sample numbers are PRJDB12479, DRA012896, and SAMD00412233, respectively.
Additional information
Funding
References
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol E. 37(1):291–294.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu ZX, Jiao BH, Nie B, Zhang GJ, Gao TG, & China Phylogeny Consortium. 2016. A comprehensive generic‐level phylogeny of the sunflower family: Implications for the systematics of Chinese Asteraceae. J Syt Evol. 54(4):416–437.
- Galbany-Casals M, Unwin M, Garcia-Jacas N, Smissen RD, Susanna A, Bayer RJ. 2014. Phylogenetic relationships in Helichrysum (Compositae: Gnaphalieae) and related genera: Incongruence between nuclear and plastid phylogenies, biogeographic and morphological patterns, and implications for generic delimitation. Taxon. 63(3):608–624.
- Kadota Y, Setoguchi H, Soejima A, Toma T, Nakata M, Morita T, Yonekura K. 2017. Asteraceae. In: Ohashi H, Kadota Y, Murata J, Yonekura K and Kihara H, editors, Wild flowers of Japan. vol. 5. Tokyo: Heibonsha; p. 198–369.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Meng Y, Sun H, Yang YP, Nie ZL. 2010. Polyploidy and new chromosome counts in Anaphalis (Asteraceae: Gnaphalieae) from the Qinghai‐Tibet Plateau of China. J Syst E. 48(1):58–64.
- Nie ZL, Funk V, Sun H, Deng T, Meng Y, Wen J. 2013. Molecular phylogeny of Anaphalis (Asteraceae, Gnaphalieae) with biogeographic implications in the Northern Hemisphere. J Plant Res. 126(1):17–32.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–11.
