Abstract
Clematis florida Thunb. is a herbaceous and perennial plant native to East Asia. The plant is resistant to cold but sensitive to heat. It is an ornamental and medicinal plant that has great commercial potential. Here, we assembled and characterized the complete chloroplast (cp) genome of C. florida. The cp genome of C. florida was characterized by Illumina pair-end sequencing and is 159,606 bp in total length. The genome includes a large single-copy (LSC) region of 79,467 bp, a small single-copy (SSC) region of 18,057 bp, and a pair of inverted repeats (IR) regions of 31,041 bp. The genome contains 135 genes including 91 protein-coding, 36 tRNA, and eight rRNA genes. Phylogenetic analysis based on 18 Clematis species indicates that C. florida is closely related to C. fusca in the Ranunculaceae. The phylogenetic relationships and taxonomic status of C. florida revealed by cp genome were consistent with the previous molecular studies, and can serve as a reference for future studies on molecular biology, evolution, and taxonomy in the genus Clematis.
The genus Clematis contains about 355 species, many of which are used to study plant modeling, flower form, colors and florescence (Wang et al. Citation2021). One of these species, Clematis florida Thunb. (First mentioned in 1784, see in http://www.iplant.cn/) is a herbaceous, perennial plant native to East Asia (Sheng et al. Citation2014). This species is resistant to cold temperature but is heat sensitive (Jiang et al. Citation2020). This species is well-known for its high ornamental value (Jiang et al. Citation2020). C. florida, also called ‘Queen of vines’, is commonly used for landscaping and floriculture, which is a popular climbing plant worldwide (Jiang et al. Citation2020). It is also a plant source of many medicinal active ingredients including antioxidant and anti-inflammatory metabolites (Jung et al. Citation2021, Wang et al. Citation2021). The cp genome has a maternal inheritance and conserved structure, and has been used to examine the developmental and phylogenetic relationships of plants (Wang et al. Citation2018). To better understand the phylogenetic position of C. florida, we assembled and analyzed the complete cp genome of C. florida using Illumina pair-end sequencing data.
Clematis florida leaves were collected from Xinyang, Henan Province, China (Xinyang Agriculture and Forestry University: 114°13′E, 32°17′N) and preserved in liquid nitrogen. Later these specimens (Bio-sample accession: SAMN20060056) were stored at −80 °C at the Horticultural Plant Biotechnology Laboratory, Xinyang Agriculture and Forestry University. A specimen was deposited at the Herbarium of the Horticultural Plant Biotechnology Laboratory, Xinyang Agriculture and Forestry University (Contact person: Jianhua Yue, [email protected]) under the voucher code CF5601. Genomic DNA was extracted by the CTAB method (Odahara et al. Citation2009). After the DNA extraction from leaf tissues, its quantification was validated using NanoDrop spectrophotometer 2000. The library construction and sequencing were performed on the Illumina High-throughput sequencing platform (NovaSeq 6000). Approximately 11.9 GB of raw data was generated with 150 bp paired-end read lengths. The data were filtered using NOVOPlasty (Dierckxsens et al. Citation2016). The complete plastid genome of C. fusca (GenBank accession: KM652489) was chosen as the reference. The plastid genome was assembled by GetOrganelle, then viewed and edited by Bandage (Wick et al. Citation2015). The cp genome annotation was performed by Geneious v 11.1.5 (Biomatters Ltd, Auckland, New Zealand) (Kearse et al. Citation2012).
The complete cp genome of C. florida is circular and 159,606 bp in length, with 37.51% GC content. The total plastid genome consisted of four distinct regions including the LSC region of 79,467 bp, SSC region of 18,057 bp, and a pair of IR regions of 31,041 bp. The complete cp genome consists of 135 genes, including 91 protein coding, 36 tRNA, and eight rRNA genes. The cp genome structure of C. florida was very close to C. fusca, which showed a cp genome of 159,609 bp in length, the LSC of 79,478 bp, SSC of 18,053 bp, and a pair of IR regions of 31,039 bp (Park and Park Citation2016).
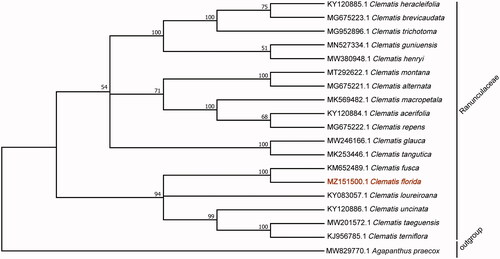
To explore the evolutionary relationship of C. florida to other species in the genus, 18 Clematis sequences were selected and Agapanthus praecox served as the outgroup for the phylogenetic analysis. All of the genomes were downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 using routine settings (Katoh and Standley Citation2013), and the phylogenetic tree was constructed by MEGA X (Kumar et al. Citation2018). The robustness of the topology was estimated using 1000 bootstrap replicates with the maximum likelihood method and nucleotide substitution model Tamura-Nei following Nguyen et al. (Citation2015). The phylogenetic tree revealed that C. florida was fully resolved in a clade with C. fusca (). The result was highly consistent with the phylogenetic relationships and taxonomic status of the Clematis species by using nuclear ITS and plastid atpB-rbcL fragments methods (Zhang et al. Citation2015). The analysis of the cp genome of C. florida provides excellent genetic information for further studies of this species and the taxonomy, phylogenetics, and evolution of the Ranunculaceae.
Author contributions
Jianhua Yue planned the research and provided the material for sequencing. Qingsong Zhu did sampling. Yan Dong analyzed data and wrote the manuscript. Jianhua Yue revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Ethics statement
Plant material collection in this study was complied with the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. See in PC RoP (cites.org). The permissions of plant material collection were not required as this is a common plant species and is abundantly available in the region of Xinyang, China.
Acknowledgments
The authors wish to thank the anonymous reviewers who provided constructive comments and critical insight on this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available at https://www.ncbi.nlm.nih.gov/. The complete cp genome has been deposited in GenBank with accession number MZ151500. And the associated Bio-project, SRA, Bio-sample numbers are PRJNA743757, SRR15041511, and SAMN20060056 respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Jiang C, Bi Y, Mo J, Zhang R, Qu M, Feng S, Essemine J. 2020. Proteome and transcriptome reveal the involvement of heat shock proteins and antioxidant system in thermotolerance of Clematis florida. Sci Rep. 10(1):8883.
- Jung J, Shin M, Jeong N, Hwang D. 2021. Antioxidant and anti-inflammatory activities of ethanol extract of Clematis trichotoma Nakai. Korean J Clin Lab Sci. 53(2):165–173.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y. 2009. Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the Moss Physcomitrella patens. Plant Cell. 21(4):1182–1194.
- Park KT, Park S. 2016. Complete chloroplast genome of Clematis fusca var. coreana (Ranunculaceae). Mitochondrial DNA A. 27(6):4056–4058.
- Sheng L, Ji K, Yu L. 2014. Karyotype analysis on 11 species of the genus Clematis. Braz J Bot. 37(4):601–608.
- Wang J, Li C, Yan C, Zhao X, Shan S. 2018. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PeerJ. 6:e5349.
- Wang R, Mao C, Jiang C, Zhang L, Peng S, Zhang Y, Feng S, Ming F. 2021. One heat shock transcription factor confers high thermal tolerance in Clematis plants. IJMS. 22(6):2900.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Zhang Y, Kong HH, Yang QE. 2015. Phylogenetic relationships and taxonomic status of the monotypic Chinese genus Anemoclema (Ranunculaceae). Plant Syst Evol. 301(5):1335–1344.