Abstract
In this study, we use a specimen from wild-caught individual to determine the complete mitochondrial genome of the Amur soft-shelled turtle (Pelodiscus maackii). The complete mitogenome of P. maackii has 16,258 bp in length and consists of 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and one control region. The arrangement of genes of P. maackii is identical with previously reported mitogenomes in the family Trionychoidea. According to our result, the ML tree for the phylogenetic reconstruction revealed that the individuals used in present study is closely related with the previously reported sequences of P. sinensis (AY962573 and MG431983) in p-distance 0.7% and 2.5%.
The genus Pelodiscus is comprised of five species worldwide. In South Korea, two species of Pelodiscus are distributed, Amur soft-shelled turtle (Pelodiscus maackii Brandt, 1858) and Chinese soft-shelled turtle (Pelodiscus sinensis Wiegmann, 1835). P. sinensis and P. maackii are commonly confused in species-level identification because of their high morphological similarities. However, there are several morphological keys to distinguish (Chang et al. Citation2012; Farkas et al. Citation2019). Generally, P. maackii is known to have longer carapace length than P. sinensis and P. maackii has a no pattern and unmarked plastron with white to straw yellow. Besides, P. sinensis has a no pattern or relatively small, faint round to oval dark markings with snow white to pinkish white (Farkas et al. Citation2019). Even though P. sinensis is known as introduced species (Chang et al. Citation2012), the definite status of Pelodiscus species distributed in Korean peninsula is still controversial. So far, previous studies on Pelodiscus species in South Korea are very insignificant, especially in genetic studies, and the study on the mitogenome of P. sinensis using specimen collected from soft-shelled turtle farm is the only research conducted (Jung et al. Citation2006). Therefore, in this study, we conduct mitogenome research on the specimen collected in the natural state to clarify the species distribution state of Korean soft-shelled turtle and to confirm the phylogenetic status with related species.
One P. maackii (specimen voucher: TOR34) sample was collected from Ogye-ri (N35.009094° E126.865442°), Naju-si, Jeollanam-do, South Korea. A frozen corpse was taken and stored at −24 °C. Total genomic DNA was extracted using Qiagen DNeasy Blood and Tissue kit (Qiagen Korea Ltd, Seoul, South Korea), following the manufacturer’s instruction. The specimen and total genomic DNA are deposited at Invasive Alien Species Research Team of National Institute of Ecology (Hae-jun Baek, [email protected]). The complete mitochondrial DNA sequence was analyzed on a Hiseq2000 platform using Next-Generation Sequencing (NGS) technique (Illumina, San Diego, CA). As a result, the complete mitogenome of P. maackii (NCBI) is comprised of a 16,258 bp nucleotide. The overall PCGs nucleotide compositions found to be 35% A, 27.6% T, 26.6% C, 10.6% G and 37.3% of GC content. Eleven out of the thirteen PCGs (ND1, ND2, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, CytB) contained ‘ATG’ as start codon. The two remaining PCGs, COX1 and ND6, contained ‘GTG’ and ‘ATA’ as start codon respectively. There were five types of stop codon found as follows: T (COX3); TAG (ND1, ND2, ND3); TAA (COX2, ATP8, ATP6, ND4L, ND4, ND5, CytB); AGA (COX1); AGG (ND6).
Figure 1. Maxium-likelihood (ML) tree based on analysis of 13 protein coding genes of 23 mitogenomes of Trionychoidea species, including P. maackii and outgroup (Carettochelys insculpta FJ862792), with GTR + I + G and bootstrapping (1000 replication).
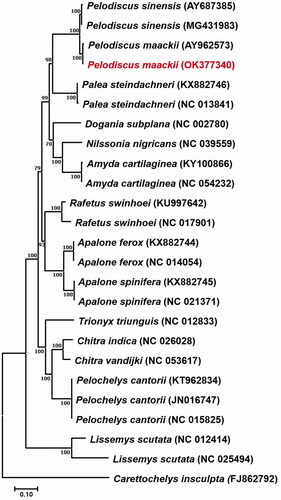
To reconstruct the phylogenetic tree for P. maackii, 14 related species in the same superfamily Trionychoidea were used and the Carettochelys insculpta (FJ862792) belonging to Carettochelyidae was used as outgroup. DNA molecular phylogeny was reconstructed with 1,000 replication of bootstrapping by maximum likelihood (ML) methodologies with MEGA 7.0 (Kumar et al. Citation2016). The best fit model was chosen on the basis of Akaike information criterion (AIC) as formed in ModelTest-NG v0.1.6 in CIPRES (Miller et al. Citation2010), and GTR + I + G model was selected. For the phylogenetic reconstruction of 14 related species in Trionychoidea was well supported (BP >70%) and performed monophyly. Our findings from the present study coincide with the results from Li et al. (Citation2017), which uses complete mitogenome of 14 species in Trionychoidea with minor difference caused by different methodologies used.
As a result, the specimen of P. maackii used in the present study is distinct, but almost identical with P. sinensis (AY962573) from the soft-shelled turtle farm in South Korea with a 0.7% of genetic distance (). The other P. sinensis (MG431983) from China showed a 2.5% of genetic difference with the specimen used. The mean interspecific divergence in the genus Pelodiscus is 2–3% (Yang et al. Citation2011; Suzuki and Hikida Citation2014), which is relatively low compared to the other genera in Trionychoidea. Since the specimen of P. sinensis (AY962573) used in the present study has been re-separated into P. maackii by Fritz et al. (Citation2010) using molecular makers and considering the finding of this research showing a similar result, it is reasonable to identify P. sinensis (AY962573) as P. maackii. This research is the first and only mitogenome study conducted on wild P. maackii in South Korea. Therefore, it is expected to make a great contribution to providing basic genetic data for the future studies of P. maackii.
Ethical consideration and permissions
The experiment was conducted in accordance with the standard operating guidelines of the animal testing and/or laboratory animal related committee (IACUC) which is published by Animal and Plant Quarantine Agency and Ministry of Food and Drug Safety of South Korea. According to the IACUC ‘Even if vertebrates are available, experiments using only animal carcasses are not subject to the deliberation category of the Institutional Animal Care and Use Committee.’ In case dead cadavers were used for present study, Ethics approval is not applicable.
Author contributions
Conception and design: Kim YC, Kim AR, Min MS, Lee H; analysis and interpretation of the data: Baek HJ, Kim PJ, Kim SH; the drafting of the paper: Baek HJ, Kim PH, Min MS; revising it critically for intellectual content; Kim PJ, Baek HJ, Lee H; the final approval of the version to be published: Baek HJ, Kim PJ. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OK377340. The associated BioProject, SRA and Bio-Sample numbers are PRJNA768941, SRX12511000 and SAMN22071900 respectively.
Additional information
Funding
References
- Chang MH, Song JY, Koo KS. 2012. The status of distribution for native freshwater turtles in Korea, with remarks on taxonomic position. Korean J Environ Biol. 30(2):151–155.
- Farkas B, Ziegler T, Pham CT, Ong AV, Fritz U. 2019. A new species of Pelodiscus from northeastern Indochina (Testudines, Trionychidae). ZK. 824:71–86.
- Fritz U, Gong S, Auer M, Kuchling G, Schneeweiß N, Hundsdörfer AK. 2010. The world’s economically most important chelonians represent a diverse species complex (Testudines: Trionychidae: Pelodiscus). Org Divers Evol. 10(3):227–242.
- Jung SO, Lee YM, Kartavtsev Y, Park IS, Kim DS, Lee JS. 2006. The complete mitochondrial genome of the Korean soft-shelled turtle Pelodiscus sinensis (Testudines, Trionychidae). DNA Seq. 17(6):471–483.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li H, Liu J, Xiong L, Zhang H, Zhou H, Yin H, Jing W, Li J, Shi Q, Wang Y, et al. 2017. Phylogenetic relationships and divergence dates of softshell turtles (Testudines: Trionychidae) inferred from complete mitochondrial genomes. J Evol Biol. 30(5):1011–1023.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, p. 1–8.
- Suzuki D, Hikid T. 2014. Taxonomic status of the soft-shell turtle populations in Japan: a molecular approach. Curr Herpetol. 33(2):171–179.
- Yang P, Tang Y, Li Ding L, Guo X, Wang Y. 2011. Validity of Pelodiscus parviformis (Testudines: Trionychidae) inferred from molecular and morphological analyses. Asian Herpetol. Res. 2(1):21–29.
