Abstract
Spiraea×vanhouttei (Rosaceae) is a frequently planted Spiraea species that is distributed in Shandong Province, Jiangsu Province, and Guangdong Province, China. The first complete chloroplast genome of Spiraea×vanhouttei was determined and described in this study. The genome is 155,957 bp in length and contained 129 encoded genes in total, including 84 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. The phylogenomic analysis showed that Spiraea×vanhouttei was closely related to Spiraea blumei according to the current sampling extent.
The genus Spiraea L. (Rosaceae) encompasses more than 100 species. Spiraea×vanhouttei (Briot) Zabel (1884) is one of Spiraea L., commonly named as Van Houtte spiraea or Vanhoutte spirea (KůdEla Citation2008). Spiraea×vanhouttei is a hybrid between Spiraea cantoniensis Lour. and Spiraea trilobata L., mainly distributed in Shandong Province, Jiangsu Province, and Guangdong Province, China (Harris Citation1917). Additionally, due to its conspicuous, luxuriant, and showy flowers, Spiraea×vanhouttei are frequently used as ornamentals and cut flowers (Yücesan et al. Citation2018). It has been used to treat diarrhea, as an astringent, anti-bacterial, and early source of salicylic acid, which is aspirin's main component. In this study, the first complete chloroplast genome of Spiraea×vanhouttei was determined and described. The study will provide potential genetic information for systematics, molecular ecology, and phylogenetic analyses in the genus Spiraea.
The leaves of Spiraea×vanhouttei were collected from Lin’an, Zhejiang, China (GPS: E119°42′44.85″, N30°16′27.04″). The specimen and extracted DNA were deposited at public herbarium of College of Life Sciences and Medicine, Zhejiang Sci-Tech University (Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, http://sky.zstu.edu.cn/, Identifier: Qiu-Ling He, [email protected]) under the voucher number ZSTULSM0002. The total genomic DNA was extracted from its silica dried leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The plastome sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA). In total, ca. 23.9 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. These clean data were de novo assembled to complete chloroplast genome using GetOrganelle (Jin et al. Citation2020), Geneious v11.1.5 (Biomatters Ltd, Auckland, New Zealand) (Drummond Citation2012) was used to annotate the genome with Spiraea insularis plastome (GenBank: MT412405) as a reference.
The full length of Spiraea×vanhouttei chloroplast sequence (GenBank accession no. MZ981785) is 155,957 bp, consisting of a large single-copy region (LSC with 84,384 bp), a small single-copy region (SSC with 18,893 bp), and two inverted repeat regions (IR with 26,340 bp). The overall GC content of Spiraea×vanhouttei chloroplast genome was 36.8% and the GC content of the LSC, SSC, and IR regions was 34.6%, 30.4%, and 42.5%. The genome contains a total of 129 genes (84 protein-coding genes, eight rRNA genes, and 37 tRNA genes). Seventeen genes had two copies, which were comprised of six PCG genes (protein-coding genes) (ndhB, rps7, ycf2, rpl2, rpl23, rps12), seven tRNA genes (trnI-CAU, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, trnL-CAA), and all four rRNA species (rrn16, rrn23, rrn4.5, rrn5). In the genome, 11 protein-coding genes (atpF, rpl2, ndhB, rps16, rpoC1, clpP, rpl16, petD, petB, rps19, ndhA) had one intron, rps12 and ycf3 gene contained two introns.
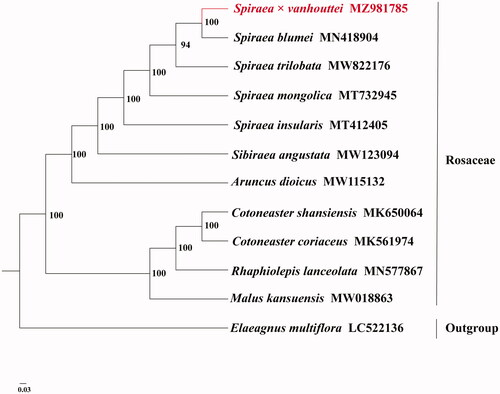
To confirm the phylogenetic position of Spiraea×vanhouttei, we obtained 10 published complete full-length chloroplast genomes of Rosaceae from NCBI. Elaeagnus multiflora was used as an outgroup for constructing the phylogenetic tree. The entire full-length chloroplast genome sequence alignment was conducted using MAFFT v7.3 (Katoh and Standley Citation2013). The phylogenetic relationship was analyzed on the complete chloroplast genomes with maximum-likelihood (ML) method using IQTREE v1.6.7 (Nguyen et al. Citation2015). The ML phylogeny was inferred under the best-selected TVM + F+R3 model and 5000 bootstrap replicates. The phylogenetic tree revealed that Spiraea×vanhouttei was closely related to Spiraea blumei with high support according to the current sampling extent ().
Authors contributions
Min-min Chen: conception and design, analysis and interpretation of the data, the drafting of the paper. Rui-hong Wang, Hong-kun Sha, and Ming-zhi Liu: data analysis. Jian-quan Tong: interpretation of data for the work and revising it critically for intellectual content. Qiu-Ling He: review, editing, and funding acquisition. All authors have agreed to be accountable for all aspects of the work.
Acknowledgements
Ethical approval: Research and collection of plant material were conducted according to the guidelines provided by Zhejiang Sci-Tech University. The plant was collected under special permission from Zhejiang Chun'an County Agricultural and Rural Development Service Center to carry out research on the species. This article does not contain any studies with endangered or protected species performed by any of the authors. Experiments were performed in accordance with the recommendations of the Ethics Committee of Zhejiang Sci-Tech University.
Health and safety: We confirm that all mandatory laboratory health and safety procedures have been complied with in conducting any experimental work reported.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the accession no. MZ981785. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA759557, SRR15686219, and SAMN21169300, respectively.
Additional information
Funding
References
- Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Harris JA. 1917. On the distribution of abnormalities in the inflorescence of Spiraea vanhouttei. Am J Bot. 4(10):624–636.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- KůdEla V. 2008. Inflorescence blast and flower bud abnormalities of Spiraea×vanhouttei and their causes. Czech J Genet Plant Breed. 43(4):135–143.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Yücesan Z, Üçler A, Oktan E, Bayraktar A, Şafak T. 2018. Weigela floribunda ve Spiraea×vanhouttei'nin elik ile üretilmesinde farkl sera ortamlar ve büyüme hormonlarnn kklenme üzerine etkileri. Orman Fak Derg. 19:1–34.