Abstract
Electric catfishes evolved the substantial electric organ that can instantly release powerful high-voltage electricity. To better study the phylogenetic position of the electric fish (Malapterurus electricus) in catfishes, in this study, we presented the complete mitochondrial genome of M. electricus assembled by the next-generation sequencing data. The mitogenome has 16,504 bp and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes, an L-strand replication origin (OL), and a control region (D-loop). The overall base composition is A 31.08%, C 27.54%, G 14.81%, and T 26.57%. Phylogenetic analysis based on 13 PCGs of 43 species from Siluriformes showed M.electricus belonging to the Malapteruridae displayed a close relationship with Siluridae. Taken together, the complete mitochondrial genome of M. electricus would be beneficial for the study of the phylogenetic relationship of Siluriformes.
Malapterurus electricus belongs to Malapterurus; Malapteruridae; Siluriformes, is mainly distributed in the freshwater basins of tropical Africa (Carl Citation2007; Diouf et al. Citation2020). The main external morphological characteristics of M. electricus include: (1) cylindrical body; (2) scaleless body surface; (3) three pairs of whiskers around its mouth; (4) without dorsal fin (Howes Citation1985; Norris Citation2002; Welzel and Schuster Citation2021). However, unlike other catfishes, electric catfishes evolved the substantial electric organ that can instantly release powerful high-voltage electricity for predation and defense (Janetzko et al. Citation1987). Although many detailed anatomy and discharge waveform of electric organ has been studied in electric catfish (Volknandt and Zimmermann Citation1986; Schikorski et al. Citation1994a, 1994b), the special genetic evolutionary relationship of M. electricus in Siluriformes analyzed by the mitochondrial genome is still scarce. In this study, we successfully assembled and annotated the complete mitochondrial genome of M. electricus using the next-generation sequencing data.
The M. electricus sample was acquired from Xiamen (118.0995 E, 24.4685 N) and stored in a refrigerator of −80 °C at School of Ecology and Environment, Northwestern Polytechnical University (Yongxin Li; [email protected]) under the voucher number 20180901DN01. The species was identified based on morphologic features and COX1 gene (GenBank accession No. EU179811.1). All procedures were approved by the Medical and Animal Experimental Ethics Committee of Northwestern Polytechnical University and followed the guidelines for the care and use of laboratory animals. The whole genome sequencing data used in this study was produced on the sequencing platform of NovaSeq 6000 (Illumina, USA). The complete mitochondrial genome of M. electricus (Genbank accession No.OL802922) was assembled by the MitoZ software (Meng et al. Citation2019) with default parameters, which has 16,504 bp in total with overall nucleotide composition of 31.08% A, 27.54% C, 14.81% G, and 26.57% T.
The annotation of the M. electricus mitochondrial genome was conducted by the MITOS Web Server (Bernt et al. Citation2013) and tRNAscan-SE Search Server (Chan and Lowe Citation2019). The gene arrangement and transcriptional orientation in M. electricus are similar to most teleosts (Luo et al. Citation2017; Prakhongcheep et al. Citation2017; Sato et al. Citation2021) with containing 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes (12S rRNA and 16S rRNA), an L-strand replication origin (OL), and a control region (D-loop). Except for COX1 used GTG as an initiation codon, the other 12 PCGs started with the conventional initiation codon of ATG. For termination codon, six PCGs (ATP6, ATP8, COX1, ND4L, ND5 and ND6) ended with TAA, three PCGs (ND1, ND2 and ND3) with TAG, and the stop codons of COX2, ND4 and CYTB are an incomplete T–, while only COX3 with TA-. The total length of 13 PCGs is 11,373 bp, which accounting for 68.91% of the complete mitogenome. Besides, all tRNAs were predicted successfully to fold into a classical cloverleaf structure except trnS-GCT, with their length ranging from 67 bp (trnC-GCA) to 75 bp (trnL-TAA). 12S rRNA was identified located between trnF-GAA and trnV-TAC with 954 bp in length, and 16S rRNA located between trnV-TAC and trnL-TAA with 1662 bp. Furthermore, the lengths of D-loop located between trnP-TGG and trnF-GAA and OL located between trnN-GTT and trnC-GCA were 866 bp and 29 bp, respectively. Compared with the mitochondrial genome which is already present in GeneBank database (https://www.ncbi.nlm.nih.gov/nuccore/AP012016.1), our mitochondrial genome annotation information is more comprehensive and detailed. Specifically, (1) we annotated 22 tRNAs in more detail, and gave all the annotation description of anti-codon; (2) two non-coding regions are successfully annotated and explained by us.
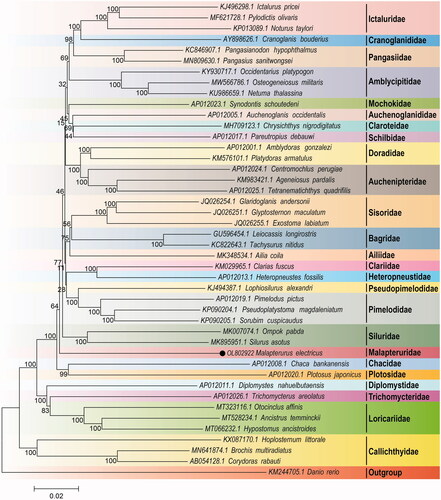
In order to systematically analyze the phylogenetic relationship of M. electricus in Siluriformes, the mitogenomes of 43 species (belongs to 25 families) from Siluriformes and one outgroup species (Danio rerio) were downloaded from GenBank nucleotide sequences database in NCBI. The multiple sequence alignment was performed by the ClustalW software (Thompson et al. Citation1994). The phylogenetic relationship was constructed based on 13 PCGs using the neighbor joining (NJ) methods with 10,000 bootstrap replications by MEGA7 (Kumar et al. Citation2016). The result of phylogenetic analysis indicated that M. electricus belonging to Malapteruridae displayed a close relationship with Siluridae (). We expect that these results would be beneficial for the study of the phylogenetic relationship of Siluriformes.
Author contributions
Y. L. conceived and supervised the project. Y. L. collected the samples and carried out the experiments. H. J. performed the bioinformatics analyses and wrote the draft manuscript. Y. L. revised the manuscript. All authors have read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The mitochondrial genome data that supports the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number OL802922. The associated BioProject, SRA, and BioSample numbers are PRJNA789410, SRR17272310, and SAMN24108793, respectively.
Additional information
Funding
References
- Bernt M, Donath A, JUhling F, Externbrink F, Florentz C, Fritzsch G, PUtz J, Middendorf M, Stadler PF. 2013. Mitos: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Carl FJ. 2007. Checklist of catfishes, recent and fossil (osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa. 1418:1–628.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Diouf K, Azeroual A, Entsua MM, Getahun A, Lalèyè P, Kazembe J. 2020. Malapterurus electricus. The IUCN red list of threatened species. e.T182850A135694315. doi:https://doi.org/10.2305/IUCN.UK.2020-2.RLTS.T182850A135694315.en
- Howes GT. 1985. The phylogenetic relationships of the electric catfish family Malapteruridae (Teleostei: Siluroidei). Annal Mag Nat Hist. 19(1):37–67.
- Janetzko A, Zimmermann H, Volknandt W. 1987. The electromotor system of the electric catfish (Malapterurus electricus): a fine-structural analysis. Cell Tissue Res. 247(3):613–624.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Luo F, Huang J, He A, Luo T, Zhou H, Wen Y. 2017. Complete mitochondrial genome of a cavefish Sinocyclocheilus ronganensis (cypriniformes: Cyprinidae). Mitochondrial DNA B Resour. 2(1):117–118.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Norris SM. 2002. A revision of the African electric catfishes, family malapteruridae (teleostei, siluriformes), with erection of a new genus and descriptions of fourteen new species, and an annotated bibliography. Ann Mus R Afr Centr Sci Zool. 289:1–155.
- Prakhongcheep O, Muangmai N, Peyachoknagul S, Srikulnath K. 2017. Complete mitochondrial genome of mouthbrooding fighting fish (Betta pi) compared with bubble nesting fighting fish (B. splendens). Mitochondrial DNA B Resour. 3(1):6–8.
- Sato M, Kawato S, Oyama H, Kaneko G, Post EJ, Suo R, Takai N, Sugita H, Kondo H, Hirono I, et al. 2021. Phylogenetic position of the Atlantic Gnomefish, Scombrops oculatus (Teleostei: Scombropidae), within the genus Scombrops, inferred from the sequences of complete mitochondrial genome and cytochrome c oxidase subunit I genes. Mitochondrial DNA B Resour. 6(10):2852–2855.
- Schikorski T, Braun N, Zimmermann H. 1994a. Immunocytochemical characterization of the synaptic innervation of a single spinal neuron, the electric catfish electromotoneuron. J Comp Neurol. 343(4):647–657.
- Schikorski T, Braun N, Zimmermann H. 1994b. Projection of brain stem neurons to the giant electromotoneurons in the cervical spinal cord of the electric catfish Malapterurus electricus. Brain Behav Evol. 43(6):306–318.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.
- Volknandt W, Zimmermann H. 1986. Acetylcholine, ATP, and proteoglycan are common to synaptic vesicles isolated from the electric organs of electric eel and electric catfish as well as from rat diaphragm. J Neurochem. 47(5):1449–1462.
- Welzel G, Schuster S. 2021. Efficient high-voltage protection in the electric catfish. J Exp Biol. 224(4):jeb239855.