Abstract
Actinodaphne lecomtei C.K.Allen, 1938 is an evergreen tree of the Lauraceae family and grows at the mountainous areas of southwestern China. In this study, we presented the first complete chloroplast genome sequence of A. lecomtei. We analyzed the chloroplast genome structure of A. lecomtei and performed a phylogenetic analysis. The complete chloroplast genome of A. lecomtei was 152,863 bp in length which contains a large single-copy (LSC) region of 93,763 bp, a small single-copy (SSC) region of 18,814 bp, and two inverted repeat (IR) regions of 20,143 bp. The analysis identified 128 genes, comprised of 84 protein-coding genes, 36 tRNAs, and eight rRNAs. The GC content of A. lecomtei complete chloroplast genome was 39.1%. The phylogenetic analysis result demonstrated that A. lecomtei was closely related to A. obovate.
Actinodaphne lecomtei C.K.Allen, 1938 is an evergreen tree of the Lauraceae family. It is mainly found in the provinces of Sichuan, Guizhou, and Yunnan in China and grows in mountains at 650–1800 m above sea level. Among the applications of A. lecomtei are making of furniture from its wood and lubricating of machines using its oil (Blanchard Citation2008; Ou et al. Citation2014). Owing to its highly conserved structure and low mutation rate, chloroplast genome has been extensively used to understand evolution and gene structure (Ebrahimi et al. Citation2021). To date, however, the chloroplast genome of A. lecomtei has not been determined. In this study, we presented the first complete chloroplast genome sequence of A. lecomtei. We analyzed the chloroplast genome structure of A. lecomtei and performed a phylogenetic analysis. This study will be useful for future studies and phylogenetic analyses of chloroplast genomes in Lauraceae species (Song et al. Citation2018).
The fresh samples of A. lecomtei were collected from Yunnan Province, China (24°23′N, 102°10′E). The study was conducted with consent from the local government and the Kunming Institute of Botany, Chinese Academy of Sciences. The voucher specimen was deposited in Qingdao University of Science and Technology (Chao Shi, [email protected]) under the specimen code AL202119. Approximately, 30 g of fresh mature leaves of A. lecomtei were used to extract chloroplast DNA using the modified high salt method previously reported (Shi et al. Citation2012). Both the quantity and quality of the extracted DNA was assessed spectrophotometrically and the integrity was assessed using 1% (w/v) agarose gel electrophoresis. DNA of high quality was sent to Novogene (Beijing, China) for genomic library construction and sequencing using the Illumina HiSeq platform (Illumina, San Diego, CA). About 4.8 Gb high quality, 2 × 150 bp pair-end raw reads were obtained and were used to assemble the complete chloroplast genome of A. lecomtei (Wang et al. Citation2018). The chloroplast genome of A. lecomtei was de novo assembled through NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) and was annotated by GeSeq (Tillich et al. Citation2017). Sequin was used to manually correct codons and gene boundaries.
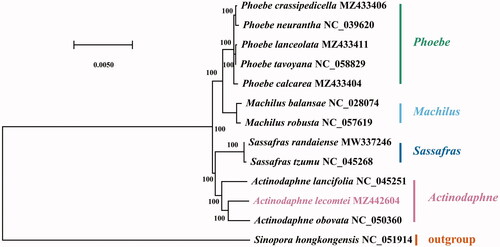
The chloroplast genome of A. lecomtei (GenBank accession MZ442604) presented a typical quadripartite structure (Wicke et al. Citation2011) with a total length of 152,863 bp which contains a large single-copy (LSC) region of 93,763 bp, a small single-copy (SSC) region of 18,814 bp, and two inverted repeat (IR) regions of 20,143 bp. The analysis identified 128 genes, comprised of 84 protein-coding genes, 36 tRNAs, and eight rRNAs. The complete chloroplast genome of A. lecomtei had the GC content of 39.1%. To reveal the evolutionary relationships between A. lecomtei and other Lauraceae, Sinopora hongkongensis was used as outgroups, together with 13 Lauraceae species to construct a phylogenetic tree (Nie et al. Citation2007; Zhang et al. Citation2021). To create sequence alignments for the construction of phylogenetic trees, MAFFT v725 (Katoh and Standley Citation2013) was used. Then the GTR-GAMMA (GTR + G) model was selected by applying the Bayesian information criterion (BIC) by Modeltest (Posada and Crandall Citation1998). Finally, MEGA-X software (Kumar et al. Citation2018) was used to perform 1000 bootstrap replications using the maximum-likelihood (ML) method. Phylogenetic analysis showed that A. lecomtei is closely related to A. obovate (). This result was similar to previous studies (Fijridiyanto and Murakami Citation2009; Song et al. Citation2017).
Authors contributions
Zimeng Chen: conceptualization, data curation, writing – original draft. Li He: methodology, validation, writing – original draft. Weicai Song: formal analysis. Wenbo Shi: software. Qin Gong: investigation. Chao Shi: resources, writing – reviewing and editing. All authors have read and approved the final manuscript.
Acknowledgements
We are thankful to Beijing-based Novogene for their NGS service that was instrumental to the execution of the project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number MZ442604. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA786749, SRR17153750, and SAMN23720001, respectively.
Additional information
Funding
References
- Blanchard OJ. 2008. Innovations in Hibiscus and Kosteletzkya (Malvaceae, Hibisceae). Novon. 18(1):4–8.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Ebrahimi A, Antonides JD, Pinchot CC, Slavicek JM, Flower CE, Woeste KE. 2021. The complete chloroplast genome sequence of American elm (Ulmus americana) and comparative genomics of related species. Tree Genet Genomes. 17(1):1–13.
- Fijridiyanto IA, Murakami N. 2009. Phylogeny of Litsea and related genera (Laureae-Lauraceae) based on analysis of Rpb2 gene sequences. J Plant Res. 122(3):283–298.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Nie ZL, Wen J, Sun H. 2007. Phylogeny and biogeography of Sassafras (Lauraceae) disjunct between Eastern Asia and Eastern North America. Plant Syst Evol. 267(1–4):191–203.
- Ou YD, Wang CB, Fan XD, Wu T. 2014. Indication effect of Guangdong indigenous broadleaf tree species to distribution pattern of climate. AMR. 1010–1012:1189–1193.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Shi C, Hu N, Huang H, Gao J, Zhao YJ, Gao LZ. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLOS One. 7(2):e31468.
- Song Y, Yao X, Liu B, Tan Y, Corlett RT. 2018. Complete plastid genome sequences of three tropical Alseodaphne trees in the family Lauraceae. Holzforschung. 72(4):337–345.
- Song Y, Yu WB, Tan Y, Liu B, Yao X, Jin J, Padmanaba M, Yang JB, Corlett RT. 2017. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the magnoliids. Genome Biol Evol. 9(9):2354–2364.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic Res. 5(1):3–13.
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297.
- Zhang Y, Tian Y, Tng DYP, Zhou J, Zhang Y, Wang Z, Li P, Wang Z. 2021. Comparative chloroplast genomics of Litsea Lam. (Lauraceae) and its phylogenetic implications. Forests. 12(6):744.