Abstract
Atriplex gmelinii C. A. Mey. Ex Bong._1838 is an annual halophytic herb found in East Asia and North America. The chloroplast genome of A. gmelinii was successfully sequenced. The assembled genome (151,852 bp; GC ratio, 37.3%) is composed of four subregions, with the large single copy (LSC; 83,872 bp; 35.4%) and small single copy (SSC; 17,812 bp; 30.9%) regions separated by two regions of inverted repeat regions (25,084 bp; 42.8%). A total of 130 genes were predicted with 85 protein-coding genes, 8 rRNAs, and 37 tRNAs. The phylogenetic analyses inferred from whole chloroplast genomes of 35 species, including 34 species in Amaranthaceae and one outgroup species, suggest a close relationship between A. gmelinii and A. centralasiatica.
Atriplex gmelinii C. A. Mey. ex Bong._1838, belonging to subfamily Chenopodioideae of family Amaranthaceae, is a halophytic annual plant found in coastal areas of Korea, China, Japan, Russia, and North America (Bassett and Crompton Citation1973; Park et al. Citation2020) and is a C4 plant (Kim et al. Citation2011). The pharmaceutical usefulness of plant extracts from A. gemelinii has been investigated by testing its antioxidant (Bae et al. Citation2003; Jeong et al. Citation2016), anticancer (Park et al. Citation2019), and anti-inflammation properties (Jeong et al. Citation2017), as well as its enhancement of osteoblast differentiation (Karadeniz et al. Citation2020). Here, we completed the chloroplast genome of A. gmelinii collected in Korea.
Total DNA of A. gmelinii collected on Apahedo Island, Shinan-gun, Jeollanam-do, Korea, (34.818343 N, 126.336961E) was extracted from fresh leaves with a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The voucher was deposited in the InfoBoss Cyber Herbarium (IN; http://herbarium.infoboss.co.kr/; Voucher number: IB-00686; Contact: Jongsun Park; [email protected]). Genome sequencing was conducted using HiSeqX at Macrogen Inc., Korea, and de novo assembly and sequence confirmation were done by Velvet v1.2.10 (Zerbino and Birney Citation2008), GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) in the Genome Information System (GeIS; http://geis.infoboss.co.kr/), which has been utilized in previous organelle genomic studies (Kwon et al. Citation2019; Oh and Park Citation2020; Park et al. Citation2020; Kim et al. Citation2021; Park et al. Citation2021). Geneious Prime® v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Chenopodium ficifolium SM_1800 chloroplast genome (NC_041200) (Kim et al. Citation2019a).
The chloroplast genome of A. gmelinii (GenBank accession: MT810472) is 151,852 bp and has four subregions: 83,872 bp of LSC and 17,812 bp of SSC regions are separated by two IR regions of 25,084 bp. It contains 130 genes (85 protein-coding genes, eight rRNAs, and 37 tRNAs); 19 genes (8 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions. The number of genes in A. gmelinii is the same as that of Atriplex centralasiatica Iljin_1936 (Zhang et al. Citation2019). However, the other Chenopodioideae chloroplast genomes showed slightly different numbers of genes: those of Chenopodium and Dysphania chloroplast genomes have 84 (Hong et al. Citation2017; Kim et al. Citation2019; Park and Kim Citation2019; Kim et al. Citation2019a, Citation2019b; Park et al. Citation2021), with the exception of Dysphania botrys (L.) Mosyakin & Clemants_2002, which has 83 genes (Chen and Yang Citation2018) and one Chenopodium album L._1753 (MF418659), which has 89 genes (Jashmi and Biseshwori Citation2017). The C. album (MF418659) may have been misidentified due to a different configuration from the rest of C. album (Park et al. Citation2021). The overall GC content is 37.3% and those in the LSC, SSC, and IR regions are 35.4%, 30.9%, and 42.8%, respectively, which is similar to those of Chenopodium species (Hong et al. Citation2017; Kim et al. Citation2019a; Park et al. Citation2021).
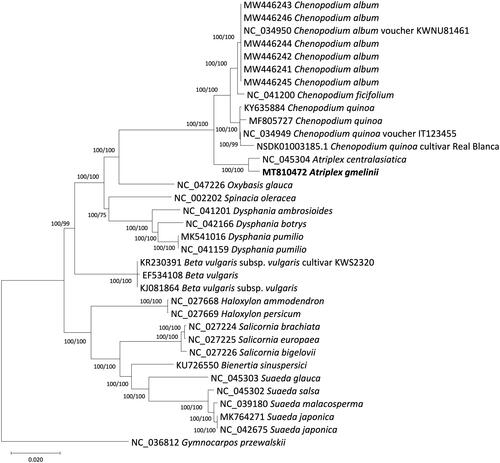
Thirty-four Chenopodioideae (Amaranthaceae) chloroplast genomes and one outgroup species, Gymnocarpos przewalski Maxim._1880 (Caryophyllaceae), were used for phylogenetic analysis. We used MEGA X (Kumar et al. Citation2018) to construct a Maximum-Likelihood (ML) tree and MrBayes v3.2.6 (Ronquist and Huelsenbeck Citation2003) to carry out Bayesian Inference (BI) after aligning whole chloroplast genomes by MAFFT v7.450 (Katoh and Standley Citation2013). A heuristic search was used with nearest-neighbor interchange branch swapping, the Tamura-Nei model, and uniform rates among sites to construct an ML phylogenetic tree with default values for other options. To estimate the node confidences, bootstrap analysis with 1,000 pseudoreplicates was conducted. For the Bayesian inferences, the GTR model with gamma rates was used as a molecular model and a Markov-Chain Monte Carlo algorithm was employed for 1,000,000 generations with four chains running simultaneously. To build the consensus tree of BI we sampled trees every 200 generations after removing 100 K generations as a ‘burn-in.’ All phylogenetic trees inferred from ML and BI methods showed that A. gmelinii is strongly grouped with A. centralasiatica (). The Atriplex group is a sister to the group composed of three species (C. album, C. ficifolium, and Chenopodium quinoa Willd._1798) in the genus Chenopodium.
Figure 1. The phylogenetic tree inferred from 34 whole chloroplast genomes of 23 species in the subfamily Chenopodioideae (Amaranthaceae) and one outgroup species in Caryophyllaceae. The ML tree is presented with the bootstrap values calculated from a ML search (1,000 bootstrap replication) and with the posterior probabilities from Bayesian inference.
Ethical statements
Authors declare that there is no ethical or legal violation in obtaining the study materials and preforming research. The species used in this study is not listed in the IUCN Red List and plant materials were collected in the location that was not designated as a protective area in Korea. Authors confirmed that the plant materials for this study were not subjected to be approved from Institutional Review Board (IRB) in the Catholic University of Korea.
Author contributions
JP and STK conceptualized the project and designed the experiment. JP, YK generated sequencing data. JP, JM and STK analyzed the data. JP and STK wrote the manuscript with input from all other authors. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Chloroplast genome sequence can be accessed via accession number of MT810472 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, BioSample, and SRA study numbers are PRJNA692662, SAMN17360090 (SRS8042657), and SRP302073(SRR13449797), respectively.
Additional information
Funding
References
- Bae J, Yoon H, Choo Y, Song S. 2003. The responses of antioxidative enzymes and salt tolerance of Atriplex gmelinii. Korean J Ecol. 26:273–280.
- Bassett I, Crompton C. 1973. The genus Atriplex (Chenopodiaceae) in Canada and Alaska. III. Three hexaploid annuals: A. subspicata, A. gmelinii, and A. alaskensis. Can J Bot. 51(10):1715–1723.
- Chen Y, Yang Z. 2018. Characterization of the complete plastome of Dysphania botrys, a candidate plant for cancer treatment. Mitochondrial DNA B Resour. 3(2):1214–1215.
- Hong S-Y, Cheon K-S, Yoo K-O, Lee H-O, Cho K-S, Suh J-T, Kim S-J, Nam J-H, Sohn H-B, Kim Y-H. 2017. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album. Front Plant Sci. 8:1696.
- Jashmi DR, Biseshwori T. 2017. Complete chloroplast genome sequence of Chenopodium album from Northeastern India. Microbiol Resour Announc. 5(47):e01150-17.
- Jeong H, Kim H, Ju E, Kong C-S, Seo Y. 2016. Antioxidant effect of the Halophyte Atriplex gmelinii. KSBB Journal. 31(4):200–207.
- Jeong H, Kim H, Ju E, Lee S-G, Kong C-S, Seo Y. 2017. Antiinflammatory activity of solvent-partitioned fractions from Atriplex gmelinii CA Mey. in LPS-Stimulated RAW264. 7 Macrophages. J Life Sci. 27(2):187–193.
- Karadeniz F, Oh JH, Im Lee J, Seo Y, Kong C-S. 2020. 5-dicaffeoyl-epi-quinic acid from Atriplex gmelinii enhances the osteoblast differentiation of bone marrow-derived human mesenchymal stromal cells via Wnt/BMP signaling and suppresses adipocyte differentiation via AMPK activation. Phytomedicine. 3:153225.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:https://doi.org/10.1093/molbev/mst010.
- Kim Y, Chung Y, Park J. 2019a. The complete chloroplast genome of Chenopodium ficifolium Sm. (Amaranthaceae). Mitochondrial DNA Part B. 4(1):872–873.
- Kim Y, Chung Y, Park J. 2019b. The complete chloroplast genome sequence of Dysphania pumilio (R. Br.) Mosyakin & Clemants (Amaranthaceae). Mitochondrial DNA Part B. 4(1):403–404.
- Kim M-H, Han M-S, Kang K-K, Na Y-E, Bang H-S. 2011. Effects of climate change on C 4 plant list and distribution in South Korea: a review. Korean J Agric Forest Meteorol. 13(3):123–139.
- Kim Y, Park J, Chung Y. 2019. Comparative analysis of chloroplast genome of Dysphania ambrosioides (L.) Mosyakin & Clemants understanding phylogenetic relationship in genus Dysphania R. Br. Korean J Plant Res. 32(6):644–668.
- Kim Y, Xi H, Park J. 2021. The complete chloroplast genome of Limonium tetragonum (Plumbaginaceae) isolated in Korea. Korean J Pl Taxon. 51(3):337–344.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:https://doi.org/10.1093/molbev/msy096.
- Kwon W, Kim Y, Park C-H, Park J. 2019. The complete chloroplast genome sequence of traditional medical herb, Plantago depressa Willd. (Plantaginaceae). Mitochondrial DNA Part B. 4(1):437–438.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Oh S-H, Park J. 2020. The complete chloroplast genome of Euscaphis japonica (Thunb.) Kanitz (Staphyleaceae) isolated in Korea. Mitochondrial DNA Part B. 5(3):3751–3753.
- Park J, An J-H, Kim Y, Kim D, Yang B-G, Kim T. 2020. Database of National Species List of Korea: the taxonomical systematics platform for managing scientific names of Korean native species. J Species Res. 9(3):233–246.
- Park J, Kim Y. 2019. The second complete chloroplast genome of Dysphania pumilio (R. Br.) mosyakin & clemants (Amranthaceae): intraspecies variation of invasive weeds. Mitochondrial DNA Part B. 4(1):1428–1429.
- Park MJ, Kim J, Kong C-S, Seo Y. 2019. Inhibition of MMP-2 and-9 by crude extracts and their solvent-partitioned fractions from the halophyte Atriplex gmelinii. Ocean Polar Res. 41(2):79–88.
- Park J, Min J, Kim Y, Chung Y. 2021. The Comparative Analyses of Six Complete Chloroplast Genomes of Morphologically Diverse Chenopodium album L.(Amaranthaceae) Collected in Korea. Int J Genomics. 2021:6643444–6643415.
- Park J, Xi H, Kim Y. 2021. The complete mitochondrial genome of Arabidopsis thaliana (Brassicaceae) isolated in Korea. Korean J Pl Taxon. 51(2):176–180.
- Park J, Xi H, Oh S-h. 2020. Comparative chloroplast genomics and phylogenetic analysis of the Viburnum dilatatum complex (Adoxaceae) in Korea. Korean J Pl Taxon. 50(1):8–16.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi:https://doi.org/10.1093/bioinformatics/btg180.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang X-J, Wang N, Zhang L-Y, Fan S-J, Qu X-J. 2019. Characterization of the complete plastome of Atriplex centralasiatica (Chenopodiaceae), an annual halophytic herb. Mitochondrial DNA B Resour. 4(2):2475–2476.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(S14):S2.
