Abstract
Emilia sonchifolia is a herb with antioxidant, anti-inflammatory, antitumor, and wound healing properties. The complete chloroplast genome (cp genome) of the genus Emilia was sequenced for the first time. The cp genome of E. sonchifolia is 151,474 bp in length. It contained a large single-copy (LSC) region (84,004 bp), and small single-copy (SSC) region (17,980 bp), and two inverted repeats (IRs, 24,745 bp). Phylogenetic analysis of 24 species was conducted. E. sonchifolia was found to be closely related to Pericallis hybrida and Dendrosenecio spp. The sequenced cp genome would be useful to understand the phylogeny and genomic studies of the genus Emilia.
Emilia sonchifolia (L.) DC. (de Candolle Citation1834), with the basionym Cacalia sonchifolia L. (Linnaeus Citation1753), belongs to tribe Senecioneae in the family Compositae. The species has various medicinal properties, including antioxidant and anti-inflammatory properties (Shylesh and Padikkala Citation1999). It is also found to have antitumor (Shylesh and Padikkala Citation2000) and wound healing properties (Smitharani et al. Citation2017). Moreover, it can be used to treat alcohol induced oxidative stress (Sophia et al. Citation2011).
Emilia Cass. is a genus with about 117 species around the world (POWO Citation2019), and is widely distributed. The genus Emilia is known to show incongruence in phylogeny relative to other lineages in tribe Senecioneae (Pelser et al. Citation2010). The genus is known to be paraphyletic or polyphyletic based on both nuclear ITS and plastid trnL-trnF phylogeny (Mapaya and Cron Citation2021).
Emilia sonchifolia is characterized by having pink or purplish tubular florets, lyrate lower leaves, and leaves usually turning purple on the lower side (Chen et al. Citation2011). The specimen was collected in the Chinese University of Hong Kong (22.419538°N, 114.208178°E). The voucher specimen with collector number T. Y. Siu 668 was deposited in the Shiu-Ying Hu Herbarium, School of Life Sciences, The Chinese University of Hong Kong (https://syhuherbarium.sls.cuhk.edu.hk/, David Tai Wai Lau, [email protected]).
Total genomic DNA of Emilia sonchifolia was extracted from 210 mg of fresh leaves using DNeasy Plant Pro Kit (Qiagen Co., Hilden, Germany) following the manufacturer’s protocol. Extracted DNA was quantified in a NanoDrop Lite (Thermo Fisher Scientific, Waltham, MA). The DNA quality was checked by visualization of the DNA using 1% agarose gel electrophoresis. Illumina 150 bp paired-end (PE) library was constructed and sequenced on the NovaSeq 6000 platform (Illumina Inc., San Diego, CA) by Novogene Bioinformatics Technology Co., Ltd. (https://en.novogene.com/, Beijing, China). Poor-quality reads (Phred score < 33) were trimmed using CLC Assembly Cell package v5.1.1 (CLC Inc., Aarhus, Denmark).
The reads were assembled into contigs using the CLC de novo assembler in CLC Assembly Cell package and SOAPdenovo v3.23 with default parameters. Gaps were filled by the Gapcloser module in SOAP package. The contigs were then aligned to the reference genome Senecio vulgaris (NC_046693.1), and assembled into complete chloroplast genome (cp genome). Gene annotation was performed on the GeSeq platform by using complete cp genomes of Senecio vulgaris (NC_046693.1) and Jacobaea vulgaris (NC_015543.1) as references. A few adjustments for protein-coding genes, start and stop codons were performed manually. The annotated genome was deposited in GenBank with the accession number MZ677242.
The cp genome of E. sonchifolia was 151,474 bp in length, containing a large single-copy (LSC) region (84,004 bp), small single-copy (SSC) region (17,980 bp), and two inverted repeats (IRs) (24,745 bp). The GC content was 37.04%. The cp genome contained 113 unique genes, including 80 protein-coding genes, 29 tRNA genes, and four rRNA genes.
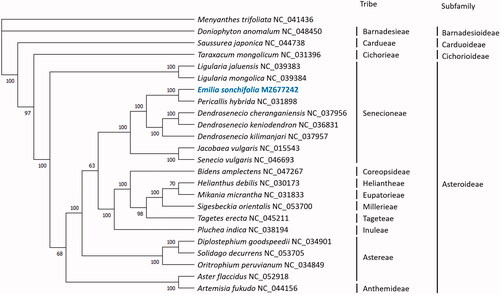
To investigate the taxonomic position of E. sonchifolia, the cp genome was aligned with 22 sequences within Compositae, and Menyanthes trifoliata (NC_041436) from the closely related family Menyanthaceae was used as an outgroup. The complete cp genomes were aligned using MAFFT7.48 (Katoh et al. Citation2019). A maximum-likelihood (ML) tree was constructed using MegaX (Kumar et al. Citation2018) based on the best fit model GTR + G and 1000 bootstrap replicates. The phylogenetic tree was labeled with family, subfamily, and tribe according to the Global Compositae Database (Compositae Working Group Citation2021) (). E. sonchifolia was found to be closely related to Pericallis hybrida, which was in agreement with the Pericallis-Emilia clade (Pelser et al. Citation2010). From the phylogenetic tree based on whole chloroplast genome, it was found that E. sonchifolia was closely related to neotropical Dendrosenecio spp., which are large alpine trees (Young and Peacock Citation1992; Knox and Palmer Citation1995). Further study is required to resolve the intergeneric relationship between the genus Emilia and other members of the tribe Senecioneae.
Authors contributions
Conceptualization: T.Y.S., D.T.W.L., P.C.S.; methodology: T.Y.S., B.L.H.K., K.H.W., H.Y.W., G.W.C.B.; data analysis: T.Y.S., B.L.H.K., K.H.W.; writing-original draft: T.Y.S.; writing-review and editing: K.H.W., B.L.H.K., H.Y.W., G.W.C.B., D.T.W.L., P.C.S.; supervision: D.T.W.L., P.C.S.
Acknowledgements
Ethical statement: This research was conducted in accordance with the Legislation of Hong Kong Special Administrative Region. The sample collections did not cause any environmental problem.
This work was supported by Wu Jieh Yee Charitable Foundation Limited.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov) with the accession number MZ677242 (https://www.ncbi.nlm.nih.gov/nuccore/MZ677242.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA796480, SRR17651893, and SAMN24864240, respectively.
References
- Chen Y, Nordenstam B, Jeffrey C. 2011. Emilia. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China Volume 20–21 (Asteraceae). Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; p. 542–543.
- Compositae Working Group. 2021. Global Compositae Database; [accessed 2021 Jul 27]. https://www.compositae.org/aphia.php?p=browser.
- de Candolle AP. 1834. Compositae Wightianae. In: Wight R, editor. Contributions to the botany of India. London: Parbury, Allen & Co.; p. 24.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Knox EB, Palmer JD. 1995. The origin of Dendrosenecio within the Senecioneae (Asteraceae) based on chloroplast DNA evidence. Am J Bot. 82(12):1567–1573.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Linnaeus C. 1753. Species Plantarum. Vol. 2. Holmiæ. Stockholm: Impensis Laurentii Salvii; p. 835.
- Mapaya RJ, Cron GV. 2021. A phylogeny of Emilia (Senecioneae, Asteraceae)–implications for generic and sectional circumscription. Taxon. 70(1):127–138.
- Pelser PB, Kennedy AH, Tepe EJ, Shidler JB, Nordenstam B, Kadereit JW, Watson LE. 2010. Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies. Am J Bot. 97(5):856–873.
- POWO. 2019. Plants of the World Online. Emilia Cass. Kew: Royal Botanic Gardens; [accessed 2021 Jul 25]. http://www.plantsoftheworldonline.org/taxon/urn:lsid:ipni.org:names:329398-2.
- Shylesh BS, Padikkala J. 1999. Antioxidant and anti-inflammatory activity of Emilia sonchifolia. Fitoterapia. 70(3):275–278.
- Shylesh BS, Padikkala J. 2000. In vitro cytotoxic and antitumor property of Emilia sonchifolia (L.) DC in mice. J Ethnopharmacol. 73(3):495–500.
- Smitharani RK, Baby B, Rasheed S, Azeem A, Kumar S. 2017. Investigation on the wound healing activity of aqueous extract of Emilia sonchifolia (L.) DC. Int J Herb Med. 5:34–39.
- Sophia D, Gomathy M, Shebin T, Ragavendran P, Arulraj C, Gopalakrishnan VK. 2011. Effect of Emilia sonchifolia (Linn.) DC on alcohol-induced oxidative stress in pancreas of male albino rats. Asian Pac J Trop Med. 4(12):973–977.
- Young TP, Peacock MM. 1992. Giant senecios and alpine vegetation of Mount Kenya. J Ecol. 80(1):41–148.