Abstract
Michelia macclurei (Dandy, 1928) is an evergreen broad-leaved tree species native to South China. This species has great ecological and economic importance. However, the genomic study of M. macclurei has lagged far behind. Here, we reported the complete chloroplast genome sequence of M. macclurei. The chloroplast genome size of M. macclurei was 160,139 bp, consisting of a pair of inverted repeat (IR) regions (26,575 bp), which was separated by a large single copy (LSC) region (88,167 bp) and a small single copy (SSC) region (18,822 bp). A total of 113 unique genes were annotated, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. The overall GC content was 39.2%. Phylogenetic analysis based on 16 whole chloroplast genome sequences of Michelia species suggested that M. macclurei and M. maudiae are sister to each other, and jointly sister to M. chapensis.
Michelia macclurei (Dandy, 1928) belonging to the family Magnoliaceae, is an evergreen broad-leaved tree species endemic to South China (Jiang et al. Citation2017). The species is a valuable multipurpose tree that can be used for landscape and urban planting, soil improvement, catchment stabilization, and biological fire prevention forest belts (Wang et al. Citation2008; Niu et al. Citation2009; Zheng Citation2013). It can be planted in monoculture or a mixture with conifers (Xia et al. Citation2016). Despite its ecological and economic importance, the genomic study of M. macclurei has lagged far behind. Nowadays, chloroplast genome sequence has been extensively used for phylogenetic, comparative, and evolutionary studies, because of its lack of recombination, small effective population size, low rates of nucleotide substitutions, and usually uniparental inheritance (Lu et al. Citation2016; Firetti et al. Citation2017). Here, we reported the chloroplast genome sequence of M. macclurei, and reconstructed its phylogenetic relationship with other Michelia species.
The fresh leaves of M. macclurei were sampled from Guangdong Academy of Forestry, Guangdong Province, China (113.45°E; 23.20°N), and under special permission from Guangdong Academy of Forestry. A voucher specimen was deposited at Guangdong Academy of Forestry (www.sinogaf.cn, contact Shujing Wei, [email protected]) under the voucher number WSJ202107001. The DNA was extracted using DNA Plantzol Reagent (Invitrogen Trading (Shanghai) Co., Ltd, Shanghai, China). DNA library construction and 150-bp paired-end sequencing were performed on the Illumina HiSeq4000 platform. The chloroplast genome was assembled using GETORGANELLE pipeline (Jin et al. Citation2020), and annotated using Geneious Prime v.2021.1.1 (http://www.geneious.com) taking Michelia chapensis (MN897730) as a reference. The annotated chloroplast genome sequence of M. macclurei was deposited in GenBank (accession number: OK046128).
The chloroplast genome sequence of M. macclurei was 160,139 bp in length and exhibited the typical quadripartite structure, consisting of a pair of inverted repeat (IR) regions of 26,575 bp, separated by a large single copy (LSC) region of 88,167 bp and a small single copy (SSC) region of 18,822 bp. The chloroplast genome encoded a total of 131 genes, of which 113 (79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes) were unique and 18 (7 protein-coding genes, 7 tRNA genes, and 4 rRNA genes) were duplicated in the IR region. A total of 16 genes were found to have introns, including 10 protein-coding genes and 6 tRNA genes. Of these genes, clpP, trnA-UGC, trnI-GAU, and ycf3 had two introns, whereas atpF, ndhA, ndhB, petB, rpl2, rpoC1, rps12, rps16, trnG-UCC, trnK-UUU, trnL-UAA, and trnV-UAC had one intron. The overall GC content was 39.2%, whereas the CC contents in the IR, LSC, and SSC were 43.2%, 37.9%, 34.3%, respectively. Comparative chloroplast genome analyses of M. macclurei and other 15 previously reported chloroplast genomes of Michelia () showed the length of these chloroplast genomes ranged from 159,819 to 160,158 bp, the GC content ranged from 39.2% to 39.3%, and they collectively contained 113 common genes (e.g. Hinsinger and Strijk Citation2017; Wang et al. Citation2019; Deng et al. Citation2020; Sima et al. Citation2020; Zhai Citation2020; Zhou et al. Citation2020; Li et al. Citation2021). These results showed the chloroplast genomes within this genus are conserved in terms of genome size, genome structure, and gene content.
Figure 1. Phylogenetic tree inferred by maximum likelihood (ML) method based on complete chloroplast genomes of 16 Michelia species with Yulania denudata as an outgroup. Numbers near the nodes represent ML bootstrap values. The phylogenetic tree based on 79 protein-coding genes is completely consistent with this topology.
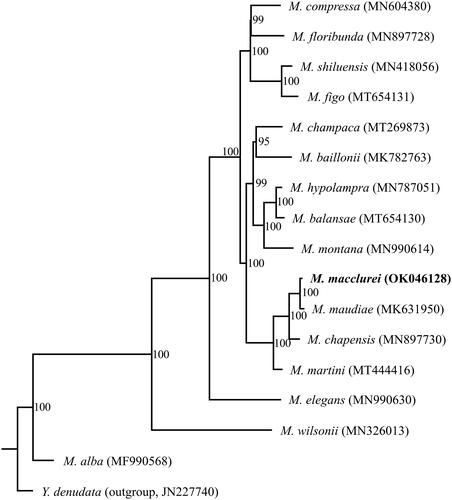
The phylogenetic relationship of Michelia was reconstructed using the maximum-likelihood (ML) method based on the multiple alignments of M. macclurei and other 15 previously reported chloroplast genomes of Michelia, with Yulania denudata (JN227740) as an outgroup. ML analysis was conducted based on two data sets: (1) the complete chloroplast genome sequences; and (2) a set of 79 common protein-coding genes, using RAxML-HPC v.8.2.8 (Stamatakis Citation2014) with 1000 bootstrap replicates on the CIPRES Science Gateway website (https://www.phylo.org/). The phylogenetic topologies based on these two data sets were completely consistent, with 100% bootstrap values at almost all nodes, and identically supported that M. macclurei and M. maudiae are sisters to each other, and jointly sister to M. chapensis ().
Authors’ contributions
WSJ conceived the project. ZYF and SZ collected samples and performed research. LSS and ZYX analyzed data. WSJ wrote the manuscript. LSS, ZYX, ZYF and SZ revised the manuscript. All authors read and approved the manuscript.
Acknowledgments
The authors are really grateful to the open raw genome data from public database.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OK046128. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA776621, SRS10788103, and SAMN22811516, respectively.
Additional information
Funding
References
- Deng Y, Luo Y, He Y, Qin X, Li C, Deng X. 2020. Complete chloroplast genome of Michelia shiluensis and a comparative analysis with four Magnoliaceae species. Forests. 11(3):267.
- Firetti F, Zuntini AR, Gaiarsa JW, Oliveira RS, Lohmann LG, Van Sluys MA. 2017. Complete chloroplast genome sequences contribute to plant species delimitation: a case study of the Anemopaegma species complex. Am J Bot. 104(10):1493–1509.
- Hinsinger DD, Strijk JS. 2017. The chloroplast genome sequence of Michelia alba (Magnoliaceae), an ornamental tree species. Mitochondrial DNA B Resour. 2(1):9–10.
- Jiang Q, Li Q, Chen Y, Zhong C, Zhang Y, Chen Z, Pinyopusarerk K, Bush D. 2017. Arbuscular mycorrhizal fungi enhanced growth of Magnolia macclurei (Dandy) figlar seedlings grown under glasshouse conditions. Forest Sci. 63(4):441–448.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Li Y, Zhou M, Wang L, Wang J. 2021. The characteristics of the chloroplast genome of the Michelia chartacea (Magnoliaceae). Mitochondrial DNA B Resour. 6(2):493–495.
- Lu RS, Li P, Qiu YX. 2016. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front Plant Sci. 7:2054.
- Niu D, Wang S, Ouyang Z. 2009. Comparisons of carbon storages in Cunninghamia lanceolata and Michelia macclurei plantations during a 22-year period in southern China. J Environ Sci. 21(6):801–805.
- Sima Y, Li Y, Yuan X, Wang Y. 2020. The complete chloroplast genome sequence of Michelia chapensis Dandy: an endangered species in China. Mitochondrial DNA B. 5(2):1594–1595.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang J, Li Y, Wang Q, Fan W. 2019. Characterization of the complete chloroplast genome of Michelia maudiae (Magnoliaceae). Mitochondrial DNA B Resour. 4(2):2146–2147.
- Wang Q, Wang S, Huang Y. 2008. Comparisons of litterfall, litter decomposition and nutrient return in a monoculture Cunninghamia lanceolata and a mixed stand in southern China. Forest Ecol Manag. 255(3–4):1210–1218.
- Xia ZC, Kong CH, Chen LC, Wang P, Wang SL. 2016. A broadleaf species enhances an autotoxic conifers growth through belowground chemical interactions. Ecology. 97(9):2283–2292.
- Zhai M. 2020. The complete chloroplast genome sequence of Michelia figo based on landscape design, and a comparative analysis with other Michelia species. Mitochondrial DNA B Resour. 5(3):2723–2724.
- Zheng BD. 2013. Analysis on the growth and fire retardant effects of Michelia macclurei in biological fire prevention forest belts at different altitudes. Anhui Forestry Sci Technol. 39:27–29.
- Zhou D, Lu S, Hou Z, Yu J. 2020. The complete chloroplast genome sequence of Michelia compressa. Mitochondrial DNA Part B. 5(3):3274–3275.
