Abstract
In this study, the complete mitochondrial genome of Steppe Whiskered Bat was sequenced for the first time using muscular tissue. The whole mitochondrial genome was 16,771 bp in length, consisting of two ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and one control region (D-loop). Phylogenetic analysis using PAUP based on mitochondrial genome (12 PCGs, except ND6) of 16 other Vespertilionidae species revealed the close relationship of M. aurascens with other related Myotis species.
Myotis aurascens (Steppe Whiskered Bat; Kuzyakin, 1935; Mammalia: Chiroptera: Vespertilionidae) distributed in south-east Mediterranean and extended eastwards out of the region into steppe Europe, and south-west Asia. Due to cryptic characteristics with M. mystacinus and M. ikonnikovi, the exact population size is not known (Benda Citation2004). In this study, the sample of M. aurascens was collected through field survey in Hulun Lake National Nature Reserve, Inner Mongolia, China (48°54′28.41″, 117°5′2.65″E). The sample was frozen in ultra-low temperature freezer and stored in the Animal Specimen Museum of Qufu Normal University, Qufu, Shandong, China (collector: Xiufeng Yang, [email protected]), and the accession number was QFA20180059. The DNA was extracted with the DNeasy Blood & Tissue kit (QIAGEN, Beverly, MA).
The sample was sequenced in an Illumina MiSeq platform with 150 PE. All sampling procedures and experimental manipulations held the proper permits (2022002). After manual assembling and annotating using online software Banklt, the genome was deposited in GenBank with the accession number OK053029.
The complete mitochondrial genome of M. aurascens is a closed circle with a length of 16,771 bp. It contains 37 genes, including two ribosomal RNA genes (rRNAs), 13 protein-coding genes (PCGs), and 22 transfer RNA genes (tRNAs), which was similar to other species of the same genus (Jebb et al. Citation2017). The light strand (L-strand) contains eight tRNA (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro) and one PCGs (ND6), and other genes are located in the heavy strand (H-strand). The overall base composition of M. aurascens was estimated to be A: 33.9%, T: 30.9%, C: 22.2%, and G: 13.0%, and the higher content of A + T (64.8%) than that of C + G (45.2%). The gene structure, content, and arrangement were found to be similar to other Myotis species reported previously (Chung et al. Citation2018).
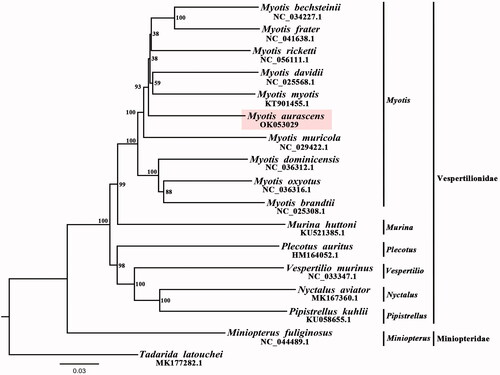
To validate the phylogenetic position of M. aurascens, 16 species (the Tadarida latouchei was chosen as an out-group) of Vespertilionidae’ 12 mitochondrial protein coding genes (except ND6) were selected to construct maximum-likelihood (ML) tree by PAUP 4.0b10 (Swofford Citation2002). According to the AIC criterion, GTR + I+G was selected as the best-fitting nucleotide substitution model using MrModeltest 3.7 (Posada Citation2005). Amino acid sequences from each PCG were aligned by MEGA11 (Koichiro et al. Citation2021). The result of Phylogenetic tree showed that M. aurascens was close to other Myotis species (). In addition, the Myotis was closely related to the Murina in the Vespertilionidae, which was also supported by previous study (Platt et al. Citation2018). We expected the data of present study to be useful for further research and phylogenetic relationship of Vespertilionidae. The data of our study are vital for the further researching Vespertilionidae and their phylogenetic relationship.
Authors contributions
Yang XF and Zhang HH conceived and designed this study; Wang Q, Bao SR, and Dou HS performed the samples collection and DNA extractions; Yang XF, Zhang L, and Zhao SH performed all bioinformatics analyses; Yang XF and Wang Q wrote the drafting of the paper; Zhang HH and Dou HS revised the critically for intellectual content and the final approval of the version to be published; and that all authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors report no conflicts of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OK053029. The associated BioProject, SRA and BioSample numbers are PRJNA768968, SRR16214749, and SAMN22072506, respectively.
Additional information
Funding
References
- Benda P. 2004. Myotis aurascens Kusjakin, 1935 – Steppen-Bartfledermaus. Wiebelsheim: Aula-Verlag; p. 1149–1158.
- Chung CU, Kim SC, Jeon YS, Han SN, Yu JN. 2018. The complete mitochondrial genome of long-tailed whiskered bat, Myotis frater (Myotis, Vespertilionidae). Mitochondrial DNA B Resour. 3(2):570–571.
- Jebb D, Foley NM, Kerth G, Teeling EC. 2017. The complete mitochondrial genome of the Bechstein's bat, Myotis bechsteinii (Chiroptera, Vespertilionidae). Mitochondrial DNA B Resour. 2(1):92–94.
- Koichiro T, Glen S, Sudhir K. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Platt RN, Faircloth BC, Sullivan KAM, Kieran TJ, Glenn TC, Vandewege MW, Lee TE, Baker RJ, Stevens RD, Ray DA. 2018. Conflicting evolutionary histories of the mitochondrial and nuclear genomes in new world myotis bats. Syst Biol. 67(2):236–249.
- Posada D. 2005. Modeltest: a tool to select the best-fit model of nucleotide substitution. Version 3.7. http://darwin.uvigo.es.
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0. Sunderland (MA): Sinauer Associates.