Abstract
The Brachionus calyciflorus species complex was recently subdivided into four species, but genetic resources to resolve phylogenetic relationships within this complex are still lacking. We provide two complete mitochondrial (mt) genomes from B. calyciflorus sensu stricto (Germany, USA) and the mt coding sequences (cds) from a German B. fernandoi. Phylogenetic analysis placed our B. calyciflorus sensu stricto strains close to the published genomes of B. calyciflorus, forming the putative sister species to B. fernandoi. Global representatives of B. calyciflorus sensu stricto (i.e. Europe, USA, and China) are genetically closer related to each other than to B. fernandoi (average pairwise nucleotide diversity 0.079 intraspecific vs. 0.254 interspecific).
Recently, the freshwater rotifer Brachionus calyciflorus Pallas, 1766 species complex was – based on genetics and morphometrics – classified into four species: Brachionus calyciflorus sensu stricto Pallas, 1766, Brachionus dorcas Gosse, 1851, Brachionus fernandoi Michaloudi, Papakostas, Stamou, Neděla, Tihlaříková, Zhang & Declerck, 2018 and Brachionus elevatus Michaloudi, Papakostas, Stamou, Neděla, Tihlaříková, Zhang & Declerck, 2018 (Papakostas et al. Citation2016; Michaloudi et al. Citation2018) which show ecological differences, such as temperature adaptation (Paraskevopoulou et al. Citation2018). Several mitochondrial (mt) genomes of Brachionus rotifers are available: B. plicatilis Muller, 1786 (Suga et al. Citation2008), B. koreanus Hwang, Dahms, Park & Lee, 2013 (Hwang et al. Citation2014), B. rotundiformis Tschugunoff, 1921 (Kim et al. Citation2017), B. rubens Ehrenberg, 1838 (Choi et al. Citation2020), B. paranguensis Guerrero-Jiménez, Vannucchi, Silva-Briano, Adabache-Ortiz, Rico-Martínez, Roberts, Neilson & Elías-Gutiérrez, 2019 (Choi et al. Citation2019), and B. angularis Gosse, 1851 (Kim et al. Citation2020). For the B. calyciflorus species complex, two mtgenomes were published so far (Nie et al. Citation2016; Choi et al. Citation2019), but without taking into account the new species delimitations.
For the newly described species of the B. calyciflorus species complex, we provide two new mtgenomes of B. calyciflorus sensu stricto from hitherto untyped regions (IGB, Germany, exact origin unknown; Oneida Lake, USA, 43°12′49.4″N 75°55′31.7″W) and coding sequences (cds) of B. fernandoi (A10, Germany, 52°30′27.2″N 13°17′14.8″E). Samples were taken from public waters and do not require a permission. Samples are permanently stored at the University of Potsdam, Germany (https://www.uni-potsdam.de, Ralph Tiedemann, [email protected]) under the voucher numbers: IGB, Oneida and A10. Strains were cultured under laboratory conditions (20 °C in a 16:8 light:dark photoperiod, details in Paraskevopoulou et al. Citation2018) for more than 10 years. Individuals were filtered through a 30 µm sieve, re-suspended in WC medium (details in Paraskevopoulou et al. Citation2018) in a 50 mL tube and centrifuged at 2000 × g for 10 min to pellet phytoplankton and other debris, before transferring the rotifers into 300 µL of TRIzol LS and storing them at –80 °C. RNA was extracted using a customized Trizol/chloroform protocol and built into an Illumina NextSeq/HiSeq library using a NEXTflex Rapid Directional RNA-Seq Library Prep kit. Sequences were analyzed on an Illumina NextSeq 500 (Oneida) or HiSeq (IGB, A10) (Novogene, Hong Kong). Raw data are permanently stored on the NCBI Short Read Archive (accession numbers SRR9040995-8 and SRR10426055-76; Paraskevopoulou et al. Citation2019, Citation2020).
Using iterative mapping, mtgenomes were reconstructed with MITObim v.1.8 (Hahn et al. Citation2013) using default parameters and a mismatch value of 3–25. We used both available B. calyciflorus mtgenomes as bait for 2–10 independent runs with 1–9 iterations per run: Netherlands (GenBank accession MN417951.1/MN417952.1) and China (KX822781.1/KX822782.1). Consensus sequences for each independent run were constructed with ANGSD v.0.95 specifying the most common base call option. Resulting consensus sequences were aligned using ClustalW (Thompson et al. Citation2002), and final consensus sequences were called using a 75% base call threshold. Automatic annotation was performed with MITOS (Bernt et al. Citation2013). The complete mtgenomes varied in length from 27,413 to 28,162 bp for chromosome I and from 9961–9988 bp for chromosome II, in line with previously published mtgenomes of B. calyciflorus. All protein-coding genes, tRNAs and rRNAs were found (GenBank accession MZ706949, MZ706950 (Germany, IGB), MZ706951, MZ706952 (USA, Oneida)). With the same approach, the 12 protein-coding genes were identified for B. fernandoi (GenBank accession only for cds MZ768793–MZ768804). These sequences were aligned with other Brachionus species from GenBank using ClustalW. A maximum-likelihood phylogenetic tree was constructed using RaxML v.8 (Stamatakis Citation2014), performing 1000 bootstrap replicates with Rotaria rotatoria (Pallas, 1766) as an outgroup.
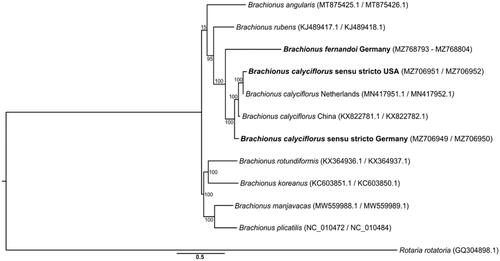
The phylogenetic analysis grouped both the German and the USA B. calyciflorus sensu stricto together with previously published B. calyciflorus specimens, suggesting that these mtgenomes derived from B. calyciflorus sensu stricto. B. fernandoi was placed with 100% bootstrap support as sister to the B. calyciflorus sensu stricto clade (). This phylogenetic grouping, together with the average pairwise nucleotide divergence, illustrates that B. calyciflorus sensu stricto lineages across the world (i.e. Europe, China, and USA) are genetically much closer to each other (0.079 ± 0.033) than to their sister species B. fernandoi (0.254 ± 0.010). Our new mtgenomes constitute a resource for future studies on the B. calyciflorus species complex and support the monophyly of the globally distributed B. calyciflorus sensu stricto.
Figure 1. Maximum-likelihood tree based on full protein coding sequences of Brachionus rotifers. Adding our new genomes (bold) highlights the monophyly of both the B. calyciflorus species complex (albeit so far only represented by two of the four species) and the B. calyciflorus sensu stricto. The numbers on branches represent percentage bootstrap support.
Authors contributions
K.K., B.D., and R.T. designed the research. S.P. processed the samples in the laboratory and analyzed the data with input from R.T. and G.W. K.K. and B.D. extracted the mitochondrial genomes. K.K. wrote the manuscript with the support of B.D., G.W., S.P., and R.T. All authors approved the final version of the manuscript.
Acknowledgements
The authors thank Dr. Michael Westbury from the GLOBE Institute, University of Copenhagen (Denmark) for his assistance with data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available on GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no MZ706949, MZ706950 (B. calyciflorus sensu stricto, Germany), MZ706951, MZ706952 (B. calyciflorus sensu stricto, USA), MZ768793–MZ768804 (B. fernandoi, Germany). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA541384, PRJNA544636, SRR9040995–SRR9040998, SRR10426055–SRR10426076, and SAMN11584680–SAMN115846803, SAMN11845726–SAMN11845747, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Choi B-S, Kim D-H, Lee J-S, Kim H-J, Hagiwara A, Lee J-S. 2020. Complete mitochondrial genome of the euryhaline monogonont rotifer Brachionus paranguensis (Rotifera, Brachionidae). Mitochondrial DNA B Resour. 5(1):502–503.
- Choi B-S, Lee YH, Hagiwara A, Lee J-S. 2019. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus calyciflorus (Rotifera, Brachionidae). Mitochondrial DNA B Resour. 4(2):3593–3595.
- Choi B-S, Lee YH, Lee J-S, Ogello EO, Kim H-J, Hagiwara A, Lee J-S. 2019. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus rubens (Rotifera, Brachionidae). Mitochondrial DNA B Resour. 5(1):5–6.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Hwang D-S, Suga K, Sakakura Y, Park HG, Hagiwara A, Rhee J-S, Lee J-S. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae). Mitochondrial DNA. 25(1):29–30.
- Kim M-S, Choi B-S, Ogello EO, Kim H-J, Hagiwara A, Lee J-S. 2020. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus angularis (Rotifera, Brachionidae). Mitochondrial DNA B Resour. 5(3):3754–3755.
- Kim H-S, Hwang D-S, Kim H-J, Sakakura Y, Hagiwara A, Lee J-S. 2017. Complete mitochondrial genome of the monogonont rotifer Brachionus rotundiformis (Rotifera, Brachionidae). Mitochondrial DNA B Resour. 2(1):39–40.
- Michaloudi E, Papakostas S, Stamou G, Neděla V, Tihlaříková E, Zhang W, Declerck SAJ. 2018. Reverse taxonomy applied to the Brachionus calyciflorus cryptic species complex: morphometric analysis confirms species delimitations revealed by molecular phylogenetic analysis and allows the (re)description of four species. PLOS One. 13(9):e0203168.
- Nie Z-J, Gu R-B, Du F-K, Shao N-L, Xu P, Xu G-C. 2016. Monogonont rotifer, Brachionus calyciflorus, possesses exceptionally large, fragmented mitogenome. PLOS One. 11(12):e0168263.
- Papakostas S, Michaloudi E, Proios K, Brehm M, Verhage L, Rota J, Peña C, Stamou G, Pritchard VL, Fontaneto D, et al. 2016. Integrative taxonomy recognizes evolutionary units despite widespread mitonuclear discordance: evidence from a rotifer cryptic species complex. Syst Biol. 65(3):508–524.
- Paraskevopoulou S, Dennis AB, Weithoff G, Hartmann S, Tiedemann R. 2019. Within species expressed genetic variability and gene expression response to different temperatures in the rotifer Brachionus calyciflorus sensu stricto. PLOS One. 14(9):e0223134.
- Paraskevopoulou S, Dennis AB, Weithoff G, Tiedemann R. 2020. Temperature-dependent life history and transcriptomic responses in heat-tolerant versus heat-sensitive Brachionus rotifers. Sci Rep. 10(1):13281.
- Paraskevopoulou S, Tiedemann R, Weithoff G. 2018. Differential response to heat stress among evolutionary lineages of an aquatic invertebrate species complex. Biol Lett. 14(11):20180498.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25(6):1129–1137.
- Thompson JD, Gibson TJ, Des Higgins G. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. DOI:https://doi.org/10.1002/0471250953.bi0203s00
