Abstract
Garra motuoensis, an endemic labeonine fish, was reported distributed in the lower Yarlung Tsangpo River drainage with little published biological information. Herein, we sequenced and characterized the complete mitochondrial genome of G. motuoensis, which was 16,806 bp in length, containing 13 PCGs, 22 tRNA genes, two rRNA genes, one light strand replication origin (OL), one control region (D-loop), and one replication region. Phylogenetic analysis based on 13 PCGs sequences revealed that G. motuoensis had a closest relationship with G. qiaojiensis. Then, both species clustered with other species of Garra, and next grouped with other genera of subfamily Labeoninae.
The genus Garra Hamilton 1822, the largest genus in the subfamily Labeoninae of family Cyprinidae, is a group of small to middle-sized benthic freshwater fishes widespread across tropical and sub-tropical waters from South China eastwards to West Africa westwards (Menon Citation1964). Species of this genus primarily inhabiting swift-flowing rivers and mountain streams, which are distinguished by the slender and subcylindrical body, crenulated rostral fold on the upper lip, and specialized mental adhesive disc on the lower lip (Chen et al. Citation2009; Nebeshwar and Vishwanath Citation2017).
In the Motuo reach of the lower Yarlung Tsangpo River drainage, there was only one Garra species ever recorded, i.e. G. kempi (Zhang et al. Citation1995; Yue Citation2000). However, based on recent ichthyological surveys, the original recorded G. kempi was revised as a new species G. tibetana (Gong et al. Citation2018a). Meanwhile, another two new species (G. motuoensis and G. yajiangensis) of this genus were described from this area (Gong et al. Citation2018b). As an endemic fish in this drainage, G. motuoensis was known only distributed in the Xigong River, a tributary of the Yarlung Tsangpo River. To date, studies on this species were quite limited except its taxonomic description.
In this study, specimen of G. motuoensis was collected from its type locality, the Xigong River (29°16′44″N, 95°14′56″E, 680 m elevation), which was deposited at the Museum of Aquatic Organisms at the Institute of Hydrobiology (IHB), Chinese Academy of Sciences under the voucher no. IHB 20161473 (contact person: Dr Huanshan Wang, [email protected]). The genomic DNA was extracted from muscle tissue using the protocol of Genomic DNA Extraction Kit (Tsingke, Beijing, China) according to the manufacturer's instruction. The genomic DNA sequences were determined through Illumina NovaSeq platform (Illumina, San Diego, CA). The mitochondrial genome was assembled using SPAdes 3.9 under the 150 paired-end reading strategy (Bankevich et al. Citation2012) with G. qiaojiensis as the reference species (Xiong et al. Citation2016). Then, all obtained genes were annotated in MITOS Web Server (Bernt et al. Citation2013).
Results showed that the complete mitogenome of G. motuoensis was a circular molecular with the length of 16,806 bp (GenBank accession no. OK375462) and contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and three non-coding regions: one light strand replication origin (OL), one control regions (D-loop), and one replication region. Among these genes, nine genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, ND6, tRNAGlu, and tRNAPro) were encoded on the minority strand, while the remaining were located at the majority strand. Most PCGs used ATN as the initiation codon except COX1 started with GTG. In addition to COX2 and ND4 with an incomplete termination codon ‘T’, the rest were encoded by the typical termination codons ‘TAR’. The lengths of single non-coding regions for OL, D-Loop and replication region were 35 bp, 876 bp, and 273 bp, respectively.
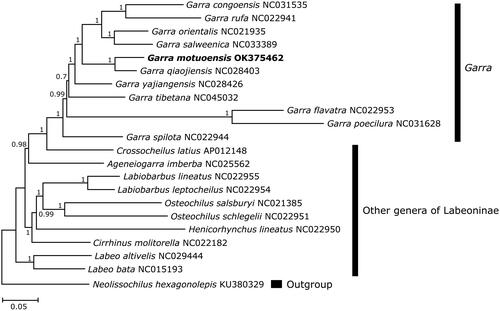
The nucleotide sequences of 13 PCGs (11,367 bp) were combined from G. motuoensis and 21 additional species including 10 species of Garra, 10 species of other labeonine genera and one outgroup (Neolissochilus hexagonolepis) for phylogenetic analysis. Sequence alignment was performed using MEGA 6.0 (Kumar et al. Citation2018). Nucleotide substitution model (GTR + I+G) was selected based on corrected Akaike information criterion (AICc) using PartitionFinder 2.1 in PhyloSuite 1.2 platform (Zhang et al. Citation2020). The phylogenetic tree was constructed based on Bayesian inference method using MrBayes 3.1 (Ronquist et al. Citation2012).
Topology of the phylogenetic tree indicated that species of genus Garra formed a highly supported monophyletic group in the subfamily Labeoninae. Specifically, G. motuoensis was a sister clade to G. qiaojiensis with the Kimura 2-parameter distance of 10.6%. Then, both species clustered with other congeners of Garra, and next grouped with species of other labeonine genera (). The present study enriched the genetic database of genus Garra, and also expanded our understanding of the phylogenetic relationships within the subfamily Labeoninae.
Figure 1. Phylogenetic tree of 21 labeonine fishes derived from Bayesian inference based on the combined nucleotide sequences of 13 protein-coding genes. The values on the nodes indicated the posterior probabilities. The GenBank accession numbers of included species were shown behind the taxon names.
Authors contributions
Zheng Gong, Xiaobing Li, and Dayong Chang were involved in the conception and design, analysis, and interpretation of the data; Zheng Gong was involved in the drafting of the manuscript; Xiaobing Li and Henglun Shen were involved in revising it critically for intellectual content and the final approval of the version to be published. All authors agree to be accountable for all aspects of this work.
Acknowledgements
Treatment procedures of animal sample have been approved by Animal Ethic Committee of Institute of Hydrobiology, Chinese Academy of Sciences, and was conducted according to ‘Instructive notions with respect to caring for laboratory animals’ issued by the Ministry of Science and Technology of the People’s Republic of China.
Disclosure statement
The authors declare that there is no potential conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession no. OK375462. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA785075, SRR17110448, and SAMN23526806, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen ZM, Zhao S, Yang JX. 2009. A new species of the genus Garra from Nujiang River Basin, Yunnan, China (Teleostei: Cyprinidae). Zool Res. 30(4):438–444.
- Gong Z, Deng SQ, Wang J, Liu HZ. 2018a. A new species of genus Garra (Cypriniformes: Cyprinidae) from the lower Yarlung Tsangpo river drainage in Southeastern Tibet, China. Chin J Zool. 53(6):857–867.
- Gong Z, Freyhof J, Wang J, Liu M, Liu F, Liu PC, Jiang YL, Liu HZ. 2018b. Two new species of Garra (Cypriniformes: Cyprinidae) from the lower Yarlung Tsangpo river drainage in southeastern Tibet, China. Zootaxa. 4532(3):367–384.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Menon AGK. 1964. Monograph of the cyprinid fishes of the genus Garra Hamilton. Mem Indian Museum. 14(4):173–260.
- Nebeshwar K, Vishwanath W. 2017. On the snout and oromandibular morphology of genus Garra, description of two new species from the Koladyne River basin in Mizoram, India, and redescription of G. manipurensis (Teleostei: Cyprinidae). Ichthyol Explor Fres. 28(1):17–53.
- Ronquist K, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Xiong H, Chen W, Gan X, Wang X. 2016. Complete sequence and rearrangement of the mitochondrial genome of Garra qiaojiensis (Cypriniformes: Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):36–37.
- Yue PQ. 2000. Fauna Sinica (Osteichthyes: Cypriniformes). Vol. III. Beijing: Science Press. (in Chinese)
- Zhang C, Cai B, Xu T. 1995. Fishes and fish resources in Xizang, China. Beijing: China Agriculture Press. (in Chinese)
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
