Abstract
This study reports the complete chloroplast genome sequence of a continental island endemic, Potentilla gageodoensis. The total plastome size was 156,397 bp, comprising one large single-copy (LSC; 85,768 bp), one small single-copy (SSC; 18,589 bp), and two inverted repeat (IR) regions (IRa and IRb, each with 26,020 bp). The overall GC content was 36.92%, and the plastome contained 131 genes, comprising 84 protein-coding genes with two pseudogenes (infA and ycf1), 37 transfer RNA genes, and eight ribosomal RNA genes. Phylogenetic analysis performed using 27 representative Rosoideae plastomes suggests that the genus Potentilla is not monophyletic and that P. gageodoensis is sister to the clade containing four taxa of Potentilla (P. freyniana, P. freyniana var. chejuensis, P. stolonifera, and P. stolonifera var. quelpaertensis). The present study reveals the taxonomic distinction of P. gageodoensis from its congeneric species in Korea and the plastome sequence obtained from this study can be used to study phylogenetic relationships and taxonomic status.
Potentilla gageodoensis M. Kim 2014, belonging to the subfamily Rosoideae of Rosaceae, is a perennial herb distributed on only a few continental islands of the Jeollanam-do Province, South Korea, in the southwestern part of the Korean Peninsula (So et al. Citation2014). It was originally described from a single island, Gageo-do Island, but was later found on Hong-do Island and a few uninhabited islands of South Korea. Potentilla L. is a large genus with approximately 400 species mainly distributed in the Northern Hemisphere (Persson et al. Citation2020). This genus has been known since ancient times, and numerous Potentilla species and their extracts have been widely used in different cultures (Tomczyk and Latté, Citation2009). Given the wide range of ploidy levels, from diploid (2x) to hexadecaploid (16x), polyploidization and hybridization have played important roles in the diversification of Potentilla (Kalkman, Citation2004; Dobes and Paule, Citation2010). Of the 17 Potentilla species known to occur in Korea, two taxa, P. dickinsii var. glabrata Nakai and P. gageodoensis, are endemic to the oceanic Ulleung Island and continental southern islands, respectively, whereas P. squamosa Sojak occurs in the central and southern Korean Peninsula (Lee, Citation2007; Heo, Lee et al. Citation2019). Morphologically, P. gageodoensis is closely related to P. fragarioides L., but it can be distinguished based on morphological characteristics including leaflet numbers, leaflet margins, and petal size (So et al. Citation2014). Little is known about the overall phylogenetic relationships among congeneric species of Potentilla in Korea as well as in East Asia. Plastome sequences of a few Potentilla species have recently been reported in Korea and China (Heo, Park et al. Citation2019; Yu et al. Citation2021). A lack of plastid genome sequences within the genus hinders our understanding of plastome organization and evolution, as well as the identification of useful plastid markers for DNA barcoding. In this study, we generated complete plastome sequences of P. gageodoensis and determined its phylogenetic position with respect to its congeneric species. This reference plastome of P. gageodoensis will be useful for future phylogenomic analyses of the genus, as well as for disentangling the complex evolutionary history of Potentilla as an effective organelle genome.
P. gageodoensis was sampled from an uninhabited island of Daesambu-do Islands (Jeollanam-do Province; N34 03.481 E127 23.71; 115 m), a small group of islands in the Jeju Strait off the southern coast of the Korean Peninsula. This sample represents an allopatric population of the original distribution of P. gageodoensis from remotely isolated southwestern islands in Korea. A voucher specimen was deposited at Sungkyunkwan University (SKK; http://115.145.139.78/bbs/board.php?bo_table=F1&wr_id=28, Seung-Chul Kim, [email protected]) under the voucher number SKK-210422503. A permit was not required given its collecting location and the specimen was identified by Seung-Chul Kim. Total genomic DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA) and sequenced using an Illumina HiSeq 4000 sequencer (Illumina, Inc., San Diego, CA, USA) at Macrogen Corporation (Seoul, Korea). A total of 51,359,878 paired-end reads were obtained and assembled de novo using Velvet v. 1.2.10 (Zerbino and Birney, Citation2008) using multiple k-mers. Transfer RNAs (tRNA) and ribosomal RNAs (rRNA) were identified using ARAGORN v.1.2.36 (Laslett and Canback, Citation2004) and RNAmmer v.1.2 Server (Lagesen et al. Citation2007), respectively. Draft annotation was conducted by transferring the annotation from the plastome of Nicotiana tabacum (NC_001879) using Geneious R v.11.1.5 (Biomatters, Auckland, New Zealand). Gene annotation was manually corrected to match the start and stop codons and intron/exon boundaries.
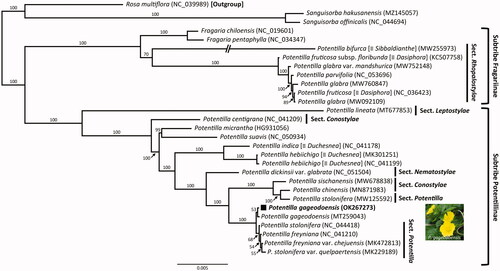
The complete plastome length of P. gageodoensis (OK267273) was 156,397 bp, comprising one large single-copy (LSC; 85,768 bp), one small single-copy (SSC; 18,589 bp), and two inverted repeat (IR) regions (IRa and IRb; 26,020 bp each). The overall GC content was 36.92%, and the plastome contained 131 genes, comprising 84 protein-coding genes with two pseudogenes (infA and ycf1), 37 tRNA genes, and eight rRNA genes. The plastome sequence of P. gageodoensis contained 17 duplicated genes in the IR regions (seven tRNA, four rRNA, and six protein-coding genes). Fifteen genes (ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained a single intron, whereas clpP and ycf3 contained two introns. The highly conserved group II intron of the atpF gene was absent in P. gageodoensis, same as in several genera of the subfamily Rosoideae (Yang et al. Citation2018; Yang, Pak et al. Citation2019; Yang, Takayama et al. Citation2019; Yang et al. Citation2020). The 27 representative plastomes of Rosoideae were aligned with default parameters using MAFFT v.7 (Katoh and Standley, Citation2013), and a maximum-likelihood (ML) analysis (the best-fit model, TVM + F + I + G4) with 1,000 ultrafast bootstrap replications was performed based on the whole plastome sequences using IQ-TREE v.1.6.7 (Nguyen et al. Citation2015). Rosa multiflora (NC_039989) was used as the outgroup. The genus Potentilla was not monophyletic; P. gageodoensis was sister to four other taxa of Potentilla, that is, P. freyniana, P. freyniana var. chejuensis, P. stolonifera, and P. stolonifera var. quelpaertensis (100% BS, ).
Author contributions
SHK and SCK involved in the conception and design of the experiments; SHK, AS, and JYY conducted analysis and interpreted the data; SHK wrote the first draft of manuscript; SCK revised the draft critically for intellectual content; SHK, AS, JYY, and SCK approved the final version and agreed to be accountable for all aspects of the work.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of this paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ (accession no. OK267273). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA766785, SRR16095409, and SAMN21876083, respectively.
Additional information
Funding
References
- Dobes C, Paule J. 2010. A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol Phylogenet Evol. 56(1):156–175.
- Heo K-I, Lee S, Kim Y, Park J, Lee S. 2019. Taxonomic studies of the tribe Potentilleae (Rosaceae) in Korea. Korean J Pl Taxon. 49(1):28–69.
- Heo K-I, Park J, Kim Y, Kwon W. 2019. The complete chloroplast genome of Potentilla stolonifera var. quelpaertensis Nakai. Mitochondrial DNA B: Resour. 4(1):1354–1356.
- Kalkman C. 2004. Potentilla. In: Kubitzki K editor. Flowering plants – Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales. Berlin: Springer, p. 366.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Lee ST. 2007. Potentilla. In: Park CW, editor. The Genera of vascular plants of Korea. Seoul: Academy Publishing Co.; p. 549–553.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Persson NL, Toresen I, Andersen HL, Smedmark JEE, Eriksson T. 2020. Detecting destabilizing species in the phylogenetic backbone of Potentilla (Rosaceae) using low-copy nuclear markers. AoB Plants. 12(3):plaa017.
- So SK, Jo H, Kim MY. 2014. A new species of Potentilla (Rosaceae): P. gageodoensis M. Kim. Korean J Pl Taxon. 44(3):175–177.
- Tomczyk M, Latté KP. 2009. Potentilla-a review of its phytochemical and pharmacological profile. J Ethnopharmacol. 122(2):184–204.
- Yang JY, Kang GH, Pak JH, Kim SC. 2020. Characterization and comparison of two complete plastomes of Rosaceae species (Potentilla dickinsii var. glabrata and Spiraea insularis) endemic to Ulleung Island. Int J Mol Sci. 21(14):4933.
- Yang JY, Pak JH, Kim SC. 2018. The complete plastome sequence of Rubus takesimensis endemic to Ulleung Island, Korea: insights into molecular evolution of anagenetically derived species in Rubus (Rosaceae). Gene. 668:221–228.
- Yang JY, Pak JH, Kim SC. 2019. Chloroplast genome of critically endangered Cotoneaster wilsonii (Rosaceae) endemic to Ulleung Island, Korea. Mitochondrial DNA B Resour. 4(2):3892–3893.
- Yang JY, Takayama K, Pak JH, Kim SC. 2019. Comparison of the whole-plastome sequence between the Bonin Islands endemic Rubus boninensis and its close relative, Rubus trifidus (Rosaceae), in the southern Korean Peninsula. Genes. 10(10):774.
- Yu S, Kang W, Yang F, Li FX. 2021. The complete chloroplast genome sequence of Potentilla glabra Lodd. Mitochondrial DNA B Resour. 6(7):1873–1874.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.