Abstract
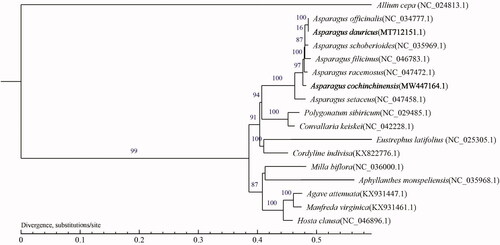
Asparagus cochinchinensis (Lour.) Merr. and Asparagus dauricus Fisch. ex Link are two traditional medical plants with therapeutic effects, distributed in mountainous regions of China. In the current study, the complete chloroplast (cp) genomes of A. cochinchinensis and A. dauricus were sequenced on the Illumina Hiseq 2500, and obtained with a length of 157,095 bp and 156,918 bp, respectively, both containing a large single-copy region and a small single-copy region separated by a pair of inverted repeat regions. The cp genome of A. cochinchinensis has 132 annotated genes including 86 protein-coding genes, 38 tRNA, and eight rRNA genes. A. dauricus has 112 annotated genes containing 78 protein genes, 30 tRNA, and four rRNA genes. The maximum-likelihood tree was reconstructed with 17 species, indicating that A. cochinchinensis is a sister group to the clade including A. officinalis to A. racemosa. This clade includes five species of Asparagus.
The Asparagus abounds with medicinal, ornamental and food plants. Asparagus cochinchinensis (Lour.) Merr., Philipp. 1919 and Asparagus dauricus Fisch. ex Link 1821 are two traditional Chinese medicine source species, which have great potential in medicine value but are difficult to distinguish morphologically (Fan et al. Citation2017). Studies on chloroplast (cp) genome will provide molecular evidence for the species identification (Kang Citation2021). Here, the cp genome of A. cochinchinensis and A. dauricus was sequenced, assembled, and analyzed with related species, and it will be used to accurately molecular identification of these species.
We collected the samples of A. dauricus and A. cochinchinensis from Nanchang (115°27′E, 28°09′N). Voucher specimen numbers (NCNU-B-1023 and NCNU-B-1024) were deposited in Botanical Specimen Museum (http://swx.ncnu.edu.cn/, the contact person is Wentao Sheng, [email protected]). The genomic DNA was extracted using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany). Sheared low molecular weight DNA fragments were used to construct paired-end libraries and sequenced on the Illumina Hiseq 2500. The raw reads were generated, retrieved and assembled using NOVOPlasty v2.6.7 (Dierckxsens et al. Citation2017). The genome annotation was performed using Geseq (Tillich et al. Citation2017) and CPGAVAS 2 (Shi et al. Citation2019). The cp genome of A. officinalis L. (NCBI accession number: NC_034777.1) was used as a reference for comparative analysis. The annotated cp genome information of A. dauricus and A. cochinchinensis was submitted to NCBI (MT712151.1 and MW447164.1).
The cp genome of A. dauricus was 156,918 bp. A pair of inverted repeats (IRa and IRb regions) was included with the length of 53,179 bp, which were separated by a large single-copy (LSC) region of 84,999 bp and a small single-copy (SSC) region of 18,740 bp. The GC content was 37.59%, and 112 unique genes were annotated, consisting of 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The cp genome of A. cochinchinensis was 157,095 bp forming a typical quadripartite structure, with an LSC region (85,306 bp), an SSC region (18,677 bp), and two IR regions (53,112 bp). The GC content was 37.48%, and 132 genes were annotated, comprising of 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes.
We downloaded the cp genomes of 17 species belonging to Asparagaceae and Allium cepa (CM022232.1) of Amaryllidaceae was selected as the out-group species to assess the relationship. The cpDNA sequences were aligned using MAFFT v7 (Katoh and Standley Citation2013), and the resulting alignments were trimmed with Gblocks (Castresana Citation2000) (get_ gblocks_ trimmed_ alignment_ from_ untrimmed.py, settings: b1 = 0.5, b2 = 0.5, b3 = 12, b4 = 7). The maximum-likelihood (ML) method was performed for the genome-wide phylogenetic analysis using PhyML 3.0 (Guindon et al. Citation2010). Nucleotide substitution model selection was estimated with jModelTest 2.1.10 (Darriba et al. Citation2012) and Smart Model Selection in PhyML 3.0. The model GTR + I+G were used for ML analyses with 1000 bootstrap replicates to calculate the bootstrap values. The result was treated with iTOL 3.4.3 (Letunic and Bork Citation2016). The evolutionary relationship indicated that A. cochinchinensis is a sister group to the clade including A. officinalis to A. racemosa (). Therefore, this study will provide important genome information for phylogenetic relationship in Asparagus.
Authors contributions
Wentao Sheng was involved in the conception and design, or analysis and interpretation of the data; the drafting of the paper, revising it critically for intellectual content; and the final approval of the version to be published; and the author agrees to be accountable for all aspects of the work.
Acknowledgements
Ethics statement: The collection of specimen conformed to the requirement of International Ethics, which did not cause damage to the local environment. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues and other conflicts of interest in this study.
Disclosure statement
We thanked Xuewen Chai for helping us identify the species of A. dauricus and A. cochinchinensis. No potential conflict of interest was reported by the author.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number MW447164.1 (A. cochinchinensis) and MT712151.1 (A. dauricus). The associated BioProject, SRA, and Bio-Sample numbers of A. cochinchinensis are PRJNA820753, SRS9706072, and SAMN20668210. The associated BioProject, SRA, and Bio-Sample numbers of A. dauricus are PRJNA752952, SRS9706070, and SAMN20668208.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fan C, Jin HZ, Wu LH, Zhang Y, Ye RD, Zhang WD, Zhang Y. 2017. An exploration of traditional Chinese medicinal plants with anti-inflammatory activities. Evid Based Complement Altern Med. 2017:1–10.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Kang Y. 2021. Molecular identification of Aquilaria species with distribution records in China using DNA barcode technology. Mitochondrial DNA B Resour. 6(4):1525–1535.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS 2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.