Abstract
The springtail Proisotoma minuta is a cosmopolitan species that can be found in many different habitats, especially in soil ecosystems. It is considered to be a good indicator of soil health. In this study, mitogenome information was obtained, which could lay a foundation for future fauna studies. The mitogenome of P. minuta is a circular module of 15,930 bp, including 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. The mitogenome of P. minuta is composed of 35.9% A, 28.5% T, 13.7% G, and 21.3% C. Phylogenetic analysis revealed that P. minuta was well grouped in the subfamily Proisotominae and had a closer relationship with Anurophorinae than Isotominae subfamily and other families.
The springtail Proisotoma minuta (Tullberg, 1871) can be found in various ecological environments and it causes damage to plant crown (Nematollahi et al. Citation2009). It has been considered an important candidate bioindicator to assess soil quality spiked with trace metals. Proisotoma minuta shows high sensitivity to cadmium, copper, and zinc, but it could tolerate high levels of lead (Nursita et al. Citation2005), arsenic (Greenslade and Vaughan Citation2003), and mercury (Buch et al. Citation2016). Proisotoma minuta may play a major role in the biological control of plant pathogens, such as Rhizoctonia solani, due to its mycophagous characteristic (Lartey et al. Citation1991). Here, the mitochondrial genome of P. minuta was assembled, which could be helpful for assessing its systematic position.
A voucher specimen of P. minuta was collected from forest soil with a humus layer of deciduous leaves at Wulian Mountain, Rizhao City, China (35.687°N, 119.395°E). The species is abundant in the field soil and does not belong to the list of key protected wild animals in China. Permission to enter Wulian Mountain for a field survey in July 2019 was acquired from the park service. After morphological identification, the collected collembolans were cultured in a laboratory environment following the standardized conditions described previously (Buch et al. Citation2016) and then stored in the laboratory of Animal Museum, School of Life Sciences, Liaocheng University (http://smkxxy.lcu.edu.cn, contact person: Xiulian Miao; email: [email protected]), under voucher no. TC202008. Total genomic DNA was extracted from the collected springtail individuals by using the CTAB method and then fragmented. The sequencing library was prepared using the Nebnext Ultra DNA Library Prep Kit for Illumina and sequenced with Illumina NovaSeq. Approximately 4.2 Gbp of clean data was obtained in this study. The mitogenome was assembled with SPAdes version 3.13.0 (Nurk et al. Citation2013) with the parameter ‘-k 127’, annotated with MITOS2 (Bernt et al. Citation2013) and deposited in GenBank with the accession number MW874475.
The complete mitogenome of P. minuta (Genbank accession no. MW874475) was sequenced to be 15,930 bp in size. The mitogenome consisted of 13 typical protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and 2 ribosomal RNA genes (rRNAs), similar to the typical mitogenome of other collembolan arthropods, such as Folsomia candida (KU198392) and Isotomurus maculatus (MK509021), but it has a different gene order compared with Pseudachorutes palmiensis (Dong et al. Citation2020). In P. minuta mitogenome, 23 genes were encoded on the H-strand, while the other 14 genes were encoded on the L-strand. The mean length of tRNAs was 64 bp, ranging from 60 to 71 bp. The overall base composition was 35.9% of A, 28.5% of T, 13.7% of G, and 21.3% of C, demonstrating a bias of higher AT content (64.4%). All PCGs started with a typical ATN codon, including seven with ATG, three with ATA, and three with ATT. Nine PCGs used TAA as the stop codon, two PCGs (atp8 and nad1) used TAG, and two (cox2 and cox3) used incomplete T.
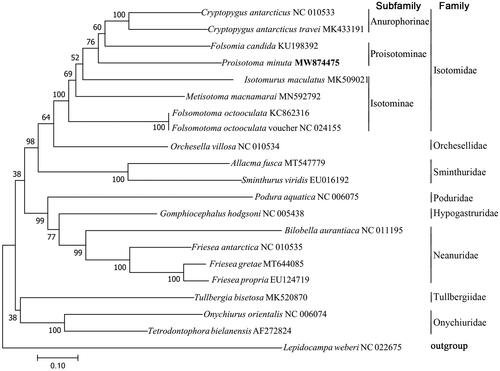
For validation of the phylogenetic position of P. minuta in Collembola, a maximum-likelihood (ML) tree was constructed on the basis of 20 Collembola mitogenome sequences, including P. minuta and one Diplura specie mitogenome sequence as an outgroup, by using MEGA 7.0 (Kumar et al. Citation2016) with the Kimura 2-parameter model and gamma-distributed with invariant sites (G + I). The robustness was tested with 2000 bootstraps. The ML tree showed that P. minuta was clustered within the subfamily Proisotominae, which is more closely related to the subfamily Anurophorinae than Isotominae subfamily and other families ().
Figure 1. Phylogenetic tree obtained from 21 mitogenome sequences. The tree was generated using maximum-likelihood analysis with Kimura 2-parameter model and gamma distributed with invariant sites (G + I). The robustness of the tree was tested with 2000 bootstraps. The numbers on the branches indicate bootstrap values.
Ethical approval
This study about the mitochondrial genome of Proisotoma minuta was granted an exemption from requiring ethical approval by the Ethics Committee of Liaocheng People’s Hospital.
Authors’ contributions
Yue Tian: springtail collection from wild fields, laboratory culture, and mitochondrial genome sequence analysis; Xiulian Miao: springtail collection and species identification; Shilin Song: auxiliary work in the laboratory and consumable material preparation; Zhengmin Zhang: springtail culture in laboratory and DNA extraction; Shiran Hu: springtail culture in laboratory and DNA extraction; Deli Wei: consumable material preparation and assistance with editing; Meng Wang: sorting out of results and paper drafting.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW874475. The associated BioProject, SRA and Bio-sample numbers are PRJNA770163, SRR16296415, and SAMN22210612, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Buch AC, Niemeyer JC, Correia MF, Silva-Filho EV. 2016. Ecotoxicity of mercury to Folsomia candida and Proisotoma minuta (Collembola: Isotomidae) in tropical soils: baseline for ecological risk assessment. Ecotoxicol Environ Saf. 127:22–29.
- Dong J, Zhang F, Wang X. 2020. Complete mitochondrial genome of Pseudachorutes palmiensis (Collembola: Neanuridae). Mitochondrial DNA B Resour. 5(1):394–395.
- Greenslade P, Vaughan GT. 2003. A comparison of Collembola species for toxicity testing of Australian soils. Pedobiologia. 47(2):171–179.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lartey R, Curl EA, Peterson C, Williams JC. 1991. Control of Rhizoctonia solani and cotton seedling disease by Laetisaria arvalis and a mycophagous insect Proisotoma minuta (Collembola). J Phytopathol. 133(2):89–98.
- Nematollahi MR, Bagheri MR, Radwanski JM. 2009. New reports of Collembola for Iran with surveying of their importance in the greenhouses of Esfahan Province. Iran Plant Prot J. 1:327–335.
- Nurk S, Bankevich A, Antipov D, Gurevich A, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology. RECOMB 2013. Vol. 7821. Berlin (Germany): Springer; p. 158–170.
- Nursita AI, Singh B, Lees E. 2005. The effects of cadmium, copper, lead, and zinc on the growth and reproduction of Proisotoma minuta Tullberg (Collembola). Ecotoxicol Environ Saf. 60(3):306–314.
