Abstract
Crepis rigescens, Diels 1921 is a traditional Chinese medicinal plant of Cichorioideae, which contains many chemicals, such as friedelin, β-sitosterol, stigmasterol, chlorogenic acid, and flavonoids, and so on, which has the characteristics of high medicinal value and small side effect. Crepis rigescens was used as folk medicines for anti-bacterial, and anti-oxidation, which also had a potential curative effect in preventing cardiovascular disease and anti-tumor. Illumina paired-end reads data were used to assemble the complete chloroplast (cp) genome. 14,425,796 raw paired-end reads and the length distribution in 124,685 bp, including a large single copy (LSC) region of 82,924 bp, a small single copy (SSC) region of 18,150 bp, and a pair of inverted repeat (IRs) regions of 25,128 bp. Besides, 10 protein-coding genes (PCGs) genes and 6 tRNAs genes possess a single intron, while clpP and ycf3 have a couple of introns. Based on the concatenated coding sequences of cp PCGs, the phylogenetic analysis showed that C. rigescens and Hypochaeris radicata (MH746729) are closely related to each other within the family Cichorioideae.
Crepis rigescens is a traditional Chinese medicinal plant of Cichorioideae, which contains many chemicals, such as friedelin, β-sitosterol, stigmasterol, Chlorogenic acid, and flavonoids, and so on, which have the characteristics of high medicinal value and small side effect (Kisiel et al. 2000; Bo et al. 2012; Ma et al. 2015). Crepis rigescens was used as folk medicines for anti-bacterial, and anti-oxidation, which also had a potential curative effect in preventing cardiovascular disease and anti-tumor (Bo et al., 2012; Tsoukalas et al. 2014).
The complete chloroplast (cp) genome consists of a pair of inverted repeats (IRs), separated by a large single-copy region (LSC) and a small single-copy region (SSC), these four parts constitute a conserved structure of the complete cp genome (Wolfe et al. Citation1992; Lee et al. Citation2007). This report will be very important for studying the phylogenetic relationships between C. rigescens and Cichorioideae.
The fresh leaves of C. rigescens were collected at Bijie, Guizhou, China (27.31 N, 105.23 E) (This plant is a common plant, and we do artificial cultivation in plants. Hence, ethical approval is not required.) and were preserved in 95% ethanol, and then transferred to a laboratory at −20 °C for long-term storage at Xi’an International University (https://www.xaiu.edu.cn/, specimen voucher: HYS 210501, Xiaoqing Liang, [email protected]). The genomic DNA was extracted with the modified CTAB method (Doyle and Doyle Citation1987). The complete Chloroplast genome sequencing and assembly were performed by Shaanxi Airui Biological Technology Co., Ltd. (Shaanxi, China). Total genomic DNA was isolated from approximately 100 mg of fresh leaves of C. rigescens using the DNeasy Plant MiniKit (Qiagen, CA, USA). After the detection of DNA purity and integrity, high-quality DNA was used for library construction and sequenced using Illumina Noveseq with a paired-end 150 strategy. Genomic DNA was used for sequencing with Illumina HiSeq X Ten Sequencing System (Illumina, CA, USA). The raw sequencing data were quality-trimmed with Geneious R11 (Biomatter Ltd., Auckland, New Zealand) and conducted with the program MITObim v 1.9 (https://github.com/chrishah/MITObim) (Hahn et al. Citation2013). The chloroplast genome of Crepidiastrum lanceolatum (MK358413) was used as the initial reference. While 4 Asteroideae were selected as outgroups (Artemisia scoparia MN385624, Galinsoga parviflora MK737938, Leucanthemum vulgare MN989913, and DTithonia diversifolia MT576958). After the assembly, the trimmed raw data were mapped to the assembled chloroplast sequence to check the assembly quality and coverage.
We assembled a 124,685 bp (GC content accounts for 37.8%) circular chloroplast genome from 14,425,796 raw paired-end reads. In addition, the length of LSC region, SSC region and IR regions distribution in 82,924 bp (GC, 36.0%), 18150 bp (GC, 31.6%), and 25,128 bp (GC, 43.0%), respectively. Based on the web-based tool OGDRaw v1.2 (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) to generate the complete cp genome (Lohse et al. Citation2013).
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession No. OM320794. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA786641, SRP349367, and SAMN23673929, respectively.
From the sequencing result, we obtained 114 complete genes, including 80 PCGs (protein-coding genes), 30 tRNAs (transfer RNAs) genes, and 4 rRNAs (ribosomal RNAs) genes. In total, 10 PCG genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps12, and rps16) harbor a single intron. 68 PCG genes have no intron. 6 tRNAs genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) harbor a single intron, and ClpP and ycf3 harbor two introns.
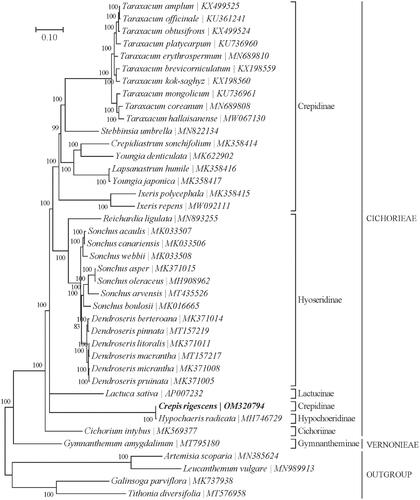
Based on the concatenated 15 cp PCGs from 36 published species of Cichorioideae and 4 Asteroideae were selected as outgroups. We constructed a Bayesian Inference (BI) phylogenetic tree () using MrBayes v3.1.2 (Milne et al. Citation2009) program integrated with TOPALi V2.5 software (Ronquist and Huelsenbeck Citation2003) to further study the phylogenetic position of C. rigescens. From the BI phylogenetic tree analysis, we find that C. rigescens and Hypochaeris radicata (MH746729) are closely related to each other within the family Cichorioideae ().
Figure 1. Phylogenetic position of Crepis rigescens based on a comparison with the complete mitochondrial genome sequences of 36 other Cichorioideae species and 4 Asteroideae as outgroups. The analysis was performed using MrBayes v3.1.2 program integrated with TOPALi V2.5 software. The accession number for each species is indicated after the scientific name.
Authors’ contributions
Peng Li’s substantial contributions to the conception or design of the work. Xiaoai Fang’s contributions are cultivation management and collection of plants. Xiaoqing Liang’s contribution is the interpretation of data for the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the Genbank database at https://www.ncbi.nlm.nih.gov/under the accession number OM320794. The associated BioProject, SRA, and BioSample numbers are PRJNA786641, SRP349367, and SAMN23673929 respectively.
Additional information
Funding
References
- Bo L, Qi YM, Zhang WZ. 2012. Study on the chemical constituents of Crepis rigescens. Journal of Qiqi har University. 28(2)22-24.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads: a baiting and iterative. Nucl Acid Res. 41(13):e129.
- Kisiel W, Zielińska K, Joshi SP. 2000. Sesquiterpenoids and phenolics from Crepis mollis. Phytochemistry. 54(8):763–766. doi:https://doi.org/10.1016/S0031-9422(00)00167-9.
- Lee HL, Jansen RK, Chumley TW, Kim KJ. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 24(5):1161–1180.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Ma SL, Li HH, Li N et al. 2015. Chemical constituents and bioactivity of Crepis tectorum linn. Journal of Qiqi Har University. 31(2):45–47.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Tsoukalas M, Gousiadou C, Skaltsa H. 2014. Guaianolides and phenolic constituents from Crepis dioscoridis L., growing wild in Greece. Phytochem. Lett. 7:202–206. doi:https://doi.org/10.1016/j.phytol.2013.11.004.
- Wolfe KH, Morden CW, Ems SC, Palmer JD. 1992. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 35(4):304–317.
