Abstract
Spinactaletes boneti mitochondrial genome was assembled and annotated. It has 14,766 bp in length, all 37 genes are present and the gene order is the same as the Pancrustacean ancestral gene order. Phylogenetic analyses based on maximum likelihood placed the species as a sister group of the remaining Entomobryomorpha, not closely related to the Isotomoidea superfamily, contradicting the actual systematics of the group.
Introduction
Spinactaletes boneti (Parisi Citation1972) is distributed along the intertidal zone of the Pacific Coast, from Zihuatanejo to Acapulco, Mexico. It belongs to the family Actaletidae (Actaletes with one species in Europe, Spinactaletes with 11 in the Americas). Representatives of this family can be distinguished from the other entomobryids by the presence of a tracheal system, abdomen III reduced, abdomen IV and V fused, abdomen VI reduced to the anal valves, abdomen IV with five sutures (two in the tergo, one intersegmental and two sternal), and 3–4 trichobothria. Spinactaletes is a genus of obligatory marine littoral springtails, mostly associated with calcareous rocks (Soto-Adames and Guillén Citation2011). The systematic position of the Actaletidae family is doubtful, some authors considered it close to Isotomidae/Isotomoidea (Yosii Citation1961; Massoud Citation1976; Soto-Adames Citation2008), to Poduridae (Paclt Citation1956; Salmon Citation1964), to Sminthuridae (Dallai & Malatesta, Citation1973), or as a sister group of all remaining Entomobryomorpha families (D’Haese Citation2003). Currently, the family is included in the Isotomoidea superfamily, because their species share a few morphological characters with some genera of Isotomoidea (Soto-Adames Citation2008).
The specimen of S. boneti sequenced here was collected from the surface of rock formations in the intertidal zone of the touristic Zihuatanejo city, Guerrero State, Mexico (17.622 N, 101.514 W) by Yun BU and José Palacios-Vargas on 10 November 2019. Twenty specimens were deposited at Shanghai Natural History Museum (Yun BU, email: [email protected]). One individual (voucher number MX-ZH-2019005) was used for DNA extraction and whole-genome amplification. All laboratory experiments including library construction and sequencing were performed by Shanghai Yaoen Biotechnology Co., Ltd, China. Illumina NovaSeq platform was used for sequencing paired-end reads with 150 bp length, producing approximately 10 G of data. The mitogenome was assembled de novo using NovoPlasty v3.8.3 (Dierckxsens et al. Citation2020) with kmer value 28 and a COI partial sequence of Mesaphorura yosii Yosii, 1906 was used as a seed (accession number KT799636.1). The identity and position of the 13 PCGs, 22 tRNA, and 2 rRNA genes were determined using MitoZ v2.4-alpha (Meng et al. Citation2019). The gene order of the new mitogenome was manually checked and it is the same as the Pancrustacean, which is the most common gene order across the Collembola group (Leo et al. Citation2019).
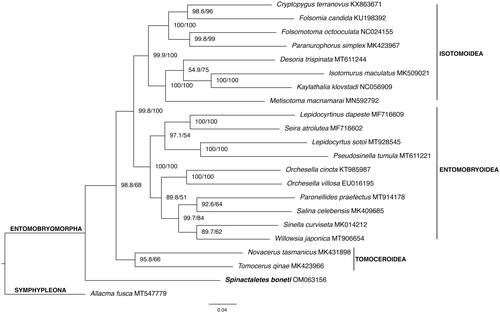
Previously to the phylogenetic analyses, mitogenomes sequences of 20 taxa of Entomobryomorpha and one of Symphypleona (outgroup) were downloaded from GenBank. All accession numbers are listed in . The newly assembled mitogenome of S. boneti (14,873 bp) was included and the final dataset comprised 22 species. Nucleotide sequences of all 13 protein coding genes were aligned using MAGUS (Smirnov and Warnow Citation2021) and BMGE v1.12 (Criscuolo and Gribaldo Citation2010) was used to trim the alignments. FASconCAT-G v1.04 (Kück and Longo Citation2014) was used to concatenate the sequences and a phylogenetic matrix with a partition scheme was created. The final matrix comprised 8084 nucleotide sites. Bayesian phylogenetic inference was performed using PhyloBayes MPI Version 1.5a (Lartillot et al. Citation2013), with CAT-GTR model, two chains were run until the likelihood had satisfactorily converged (maxdiff< 0.1). Maximum Likelihood inference was performed using IQ-Tree v2.0.7 (Minh et al. Citation2020), ultrafast bootstrap 1000 replicates (Hoang et al. Citation2018), and SH-aLRT support. Model Finder (Subha Kalyaanamoorthy et al. Citation2017) selected the best partition scheme and GTR + F substitution model for the six partitions. The phylogenetic tree was visualized and edited in FigTree v1.4.2 (available on https://tree.bio.ed.ac.uk/software/figtree/). The resulting topology based on ML () suggested the position of Spinataletes boneti as a sister group of the remaining Entomobryomorpha species with moderate bootstrap support (68%). Bayesian analyses placed the new mitogenome in the same position but with a bit lower posterior probability support (0.63). Our result is in agreement with the study made by D’Haese (Citation2003), based on 131 morphological characters from 67 Collembola taxa, which concluded that the springtails had a terrestrial edaphic origin with the semi-aquatic life representing a secondary specialization, not a primitive condition. Further analyses including more mitogenomes of Actaletidae and Coenaletidae taxa need to be carried out to verify our result, considering that the support value is not high enough to make a conclusion. Internal relationships of the superfamilies Entomobryoidea, Tomoceroidea, and Isotomoidea are not the focus of our study, so they are not discussed here.
Ethics statement
Permit to collected Spinactaletes boneti specimens in Mexico was SEMARNAT: SGPA/DGVS/04902/19, given to our colleague Dr. Margarita Ojeda. The field studies did not involve endangered or protected species. The sequenced species is common in Mexico, and is not included in the “List of Protected Animals in Mexico”.
Author contributions
Y.B. and J.P. collected the specimen, concepted and designed the idea; N.N.G. analyzed and interpretated the data; Y.B., Y.G., J.P. and N.N.G. were responsible for the drafting of the paper, for revising it and for the final approval; all authors agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OM063156. The associated **BioProject**, **SRA**, and **Bio-Sample** numbers are PRJNA792924, SRR17381101, and SAMN24475446 respectively.
Additional information
Funding
References
- Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210.
- Dallai R, Malatesta E. 1973. Ricerche sui Collemboli. XIX. La fine struttura epicuticulare di Podura ed Actaletes. Redia. 54:135–139.
- D’Haese CA. 2003. Morphological appraisal of Collembola phylogeny with special emphasis on Poduromorpha and a test of the aquatic origin hypothesis. Zoologica Scripta. 32:563–586.
- Dierckxsens N, Mardulyn P, Smits G. 2020. Unraveling heteroplasmy patterns with NOVOPlasty. NAR Genom Bioinform. 2(1):lqz011.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81.
- Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615.
- Leo C, Carapelli A, Cicconardi F, Frati F, Nardi F. 2019. Mitochondrial genome diversity in collembola: phylogeny, dating and gene order. Diversity. 11(9):169–120.
- Massoud Z. 1976. Essai de synthèse sur la phylogénie des Collemboles. Revue D’Ecologie et de Biologie du Sol. 13(1):241–252.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Research. 47(11):e63–7.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Paclt J. 1956. Biologie der Primär Flügellosen Insekten. Jena: Fischer.
- Parisi V. 1972. Actaletes boneti, a new species of Actaletidae (Collembola) of the Mexican coast of the Pacific. Accademia Naz Lincei Roma. 171:43–46.
- Salmon JT. 1964. An index to the Collembola. I. R Soc New Zealand Bull. 7:1–144.
- Smirnov V, Warnow T. 2021. MAGUS: multiple sequence alignment using graph clustering. Bioinformatics. 37(12):1666–1672.
- Soto-Adames FN, Barra J-A, Christiansen KA, Jordana R. 2008. Suprageneric classification of collembola entomobryomorpha. Ann Entomol Soc Am. 101(3):501–513.
- Soto-Adames FN, Guillén C. 2011. Two new species of the marine littoral springtail genus Spinactaletes (Collembola: Actaletidae) from Costa Rica. BMS. 87(3):463–483.
- Yosii R. 1961. Phylogenetische bedeuntung der chaetotaxiee bei den Collembolen. Contributions from the Biological Laboratory, Kyoto University, Kyoto, 12, p. 1–37.