Abstract
Peltoboykinia tellimoides, which is distributed in Japan and China, is the type species of Peltoboykinia. In this study, we sequenced and assembled the complete chloroplast (cp) genome of P. tellimoides and reconstructed the phylogeny of Saxifragaceae based on the whole cp genome sequences. The cp genome of P. tellimoides was 156,274 bp in length, comprising a pair of inverted repeat regions (25,099 bp) separated by a large single copy region (88,109 bp) and a small single copy region (17,967 bp). The genome encoded 112 unique genes consisting of 78 different protein-coding genes, 30 transfer RNA and four ribosomal RNA genes, with 16 duplicated genes in the inverted repeats. Phylogenetic analysis indicated that P. tellimoides together with three Chrysosplenium species formed a high support clade, which was sister to Micranthes melanocentra.
Peltoboykinia (Engl.) Hara is a small genus of flowering plants belonging to the family Saxifragaceae and its native range is Southeastern China and Japan (Wu and Raven Citation2001). There are only two species within the genus, P. tellimoides (Maxim.) H. Hara Citation1937 and P. watanabei (Yatabe) H. Hara Citation1937, which are characterized by yellowish dentate petals 5 clothed with glandular, stout thick rhizome, glossy peltate leaves, inflorescentia cymoso-paniculata and hypanthium campanulatus (Hara Citation1937). P. tellimoides is the type species of this genus and the rhizome of the plant contained bergenin which is often found in Saxifragaceae (Nagai et al. Citation1969). However, the lack of research on the genetic background of P. tellimoides greatly affects the utilization of this species. Here, we generated genome skimming data for P. tellimoides to assembly the cp genome. The genome sequence of P. tellimoides was deposited into GenBank with the accession number MZ779205.
One P. tellimoides individual was collected from Suichang County, Zhejiang Province of China (118°52′56.69″E, 28°21′21.79″N). A specimen was deposited at the Herbarium of Zhejiang University (HZU; Pan Li, [email protected]) under the voucher number XXL170002-1, and no specific permissions were required for sample collection and the field study did not involve Endangered or protected species. Silica-dried leaf tissues were used to extract total genomic DNA by using Plant DNAzol Reagent (LifeFeng, Shanghai) according to the manufacturer’s protocol. The high-quality DNA was sheared (yielding ≤800 bp fragments) and the short-insert paired-end libraries preparation and sequencing were performed on Illumina X10 (Beijing Genomics Institute, Wuhan, China) with read length of 150 bp. The raw data was filtered by quality with Phred score <30 and all remaining sequences were assembled into contigs using the CLC de novo assembler beta 4.06 (CLC Inc., Rarhus, Denmark). Then the software Geneious R11 (Biomatters, Auckland, New Zealand) was used to reconstruct and annotated the cp genome with Tiarella polyphylla D. Don 1825 (GenBank accession number: MH708568) as a reference according to the description in Liu et al. (Citation2017) and Liu et al. (Citation2020). Maximum likelihood (ML) method was applied to reconstruct the phylogenetic tree for 22 whole cp genome sequences of Saxifragaceae species, with two Grossulariaceae species as outgroup. ML analysis was implemented in RAxML-HPC v8.2.12 on the CIPRES cluster (Miller et al. Citation2010) using the best-fit nucleotide substitution model (GTR + I + G) determined from jModelTest v2.1.4 (Posada Citation2008), and 1000 bootstrap iterations were conducted with other parameters using the default settings.
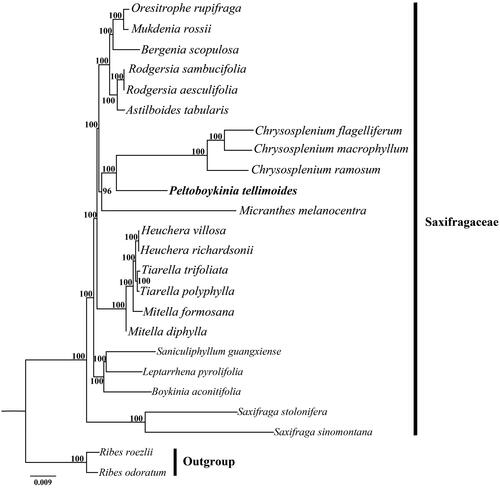
There were 35,589,288 paired-end reads generated from genome skimming sequencing for P. tellimoides, and 8,168,458 reads were removed from the raw data after trimming low quality sequences (Phred scores of 30 or less). The complete cp genome of P. tellimoides was 156,274 bp in length and shared the common feature of comprising two copies of IR (25,099 bp each) that divided the genome into two single-copy regions (LSC 88,109 bp; SSC 17,967 bp). The overall GC content of the total length, LSC, SSC, and IR regions was 37.9, 36.0, 32.7, and 43.0% respectively. Within the cp genome of P. tellimoides there were 112 unique genes, including 78 protein-coding genes, 30 tRNA genes, 4rRNA genes and 16 duplicated genes. Among them, six tRNA genes and nine protein coding genes contained a single intron, and clpP and ycf3 contained two introns. The phylogeny showed that P. tellimoides and three Chrysosplenium species, including C. flagelliferum F. Schmidt 1868, C. macrophyllum Oliv. 1888, and C. ramosum Maxim. 1859, formed a highly supported clade, which in turn was sister to Micranthes melanocentra (Franch.) Losinsk. 1896 ().
Figure 1. Phylogenetic tree reconstruction of 22 taxa of Saxifragaceae and two outgroups using ML method. Relative branch lengths are indicated at the top-left corner. Numbers above the branches represent bootstrap values from maximum-likelihood analyses. GenBank accession numbers of taxa are shown below, Oresitrophe rupifraga (MF774190), Mukdenia rossii (MG470844), Bergenia scopulosa (KY412195), Rodgersia sambucifolia (MN496077), Rodgersia aesculifolia (MW327540), Astilboides tabularis (MT316511), Chrysosplenium flagelliferum (MN729584), Chrysosplenium macrophyllum (MK973001), Chrysosplenium ramosum (MK973002), Peltoboykinia tellimoides (MZ779205), Micranthes melanocentra (MT740256), Heuchera villosa (MH708563), Heuchera richardsonii (MH708562), Tiarella trifoliata (MH708572), Tiarella polyphylla (MH708568), Mitella formosana (MH708565), Mitella diphylla (MH708564), Saniculiphyllum guangxiense (MN496078), Leptarrhena pyrolifolia (MN496070), Boykinia aconitifolia (MN496058), Saxifraga stolonifera (MH191389), Saxifraga sinomontana (MN104589), Ribes roezlii (MN496076), Ribes odoratum (MT081309).
Author contributions
MFD and LXL designed the study. RRY and RL conducted the field sampling. RRY produced and analyzed the data. MFD wrote the manuscript. LXL revised the manuscript. All authors approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ779205. The associated Bio-Project, SRA and Bio-Sample numbers of the raw sequence data for assembling the cp genome are PRJNA754512, SRR15460330, and SAMN20792568, respectively.
Additional information
Funding
References
- Hara H. 1937. Two new genera of Saxifragaceae in Japan. Shokubutsugaku Zasshi. 51(605):250–253.
- Liu LX, Du YX, Folk RA, Wang SY, Soltis DE, Shang FD, Li P. 2020. Plastome evolution in Saxifragaceae and multiple plastid capture events involving Heuchera and Tiarella. Front Plant Sci. 11:361.
- Liu LX, Li R, James RP, Worth Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010. IEEE, p. 1–8.
- Nagai M, Izawa K, Inoue T. 1969. Studies on the constituents of Saxifragaceous plants. I. Two triterpene acids of Peltoboykinia tellimoides (MAXIM.) HARA. Chem Pharm Bull. 17(7):1438–1443.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Wu Z, Raven P. 2001. Flora of China, Vol. 8: Brassicaceae Through Saxifragaceae. St. Louis, MI: Missouri Botanical Garden Press, p. 506.
