Abstract
We sequenced and annotated the complete mitochondrial genome (mitogenome) of Takydromus kuehnei Van Denburgh, 1909 (Squamata: Takydromus). This mitogenome was 17,224 bp long and encoded 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, one non-coding regions of an L-strand replication origin and a displacement loop region. The overall nucleotide composition was 32.8% of A, 13.8% of G, 24.8% of T, and 30.5% of C. Phylogenetic analysis using maximum likelihood method validated the taxonomic status of T. kuehnei, exhibiting the close relationship with the species from the genus Takydromus.
Takydromus kuehnei Van Denburgh, 1909 (Squamata: Takydromus) is a small lizard from family Lacertidae genera Takydromus and lives in secondary forest and grass, which was firstly found in Taiwan, China, and recently it was found in Wuyi mountain in Fujian, China and Zhejiang, China (Uetz et al. Citation2021). In this study, we sequenced the mitochondrial genome (mitogenome) of T. kuehnei (GenBank accession No. MZ435950).
The specimens of male T. kuehnei used in this study was collected in Quzhou, Zhejiang, China (29.45 N, 118.82 E) and its tail tissue was immediately stored in 75% ethanol at −20 °C. Then the lizard was released to sample site. The experimental procedures followed in this study complied with the animal welfare and research laws in China and were approved by the Ethics Committee of Lishui University (premit No. AREC-LSU202104001DGH). We used the Wizard® Genomic DNA Purification Kit (Promega, Madison, USA) to isolate the whole genomic DNA from tail tissues of each specimen according to the manufacturer’s instructions. The extracted genomic DNA was stored in the Laboratory of Amphibian Diversity Investigation (contact person: Guo-Hua Ding, E-mail: [email protected]) at Lishui University, and the Accession Number of the specimen was LSU20210413QLG001. The genomic DNA was sequenced using the Hiseq6000 platform (Illumina Inc., San Diego, CA, USA). The mitogenome of Takydromus amurensis (GenBank accession No. KU641018) was employed as the reference sequence (Park et al. Citation2016). We used the NOVO Plasty 3.7 (Dierckxsens et al. Citation2017) to assemble and MITOS Web Serve to annotate the mitogenome (Bernt et al. Citation2013).
We obtained the mitochondrial genome of T. kuehnei with 17,224 bp sequence. This mitogenome encoded 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs (12S and 16S) and one non-coding regions of an L-strand replication origin and a displacement loop region (control region). The overall nucleotide composition was 32.8% of A, 13.8% of G, 24.8% of T, and 30.5% of C. The gene arrangement pattern and transcription directions were identical to previous studies in the suborder Sauria (Kim et al. Citation2016). Among the 13 PCGs, the ATP8 was with the least nucleotides, while the ND5 was with the most nucleotides. All PCGs initiated with ATG as a start codon, except for APT6 gene, which began with GTG. Ten genes (ND1, ND2, ND3, COI, ATP8, ATP6, ND4L, ND5, ND6, and Cytb) ended with complete stop codons (TAA, AGA, and AGG), and the other genes ended with T or TA as the incomplete stop codons, which were presumably completed as TAA by post transcriptional polyadenylation (Anderson et al. Citation1981).
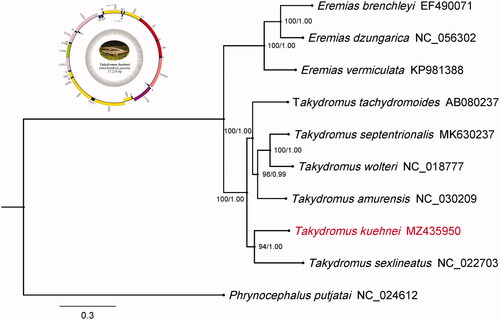
To validate the phylogenetic position of T. kuehnei, the phylogenetic tree was constructed using 13 mitochondrial PCGs, including six Takydromus species, and Eremias vermiculata, Eremias dzungarica, Eremias brenchleyi, and Phrynocephalus putjatai were used as out-group. The Bayesian Inference (BI) tree was constructed using MrBayes 3.2.6 under GTR + I + G + F (2,000,000 generations) with the initial 25% of sampled data discarded as burn-in, and maximum likelihood (ML) tree was constructed using IQ-TREE under the GTR + I+G4 + F model for 5000 ultrafast bootstraps in PhyloSuite v1.2.2 (Zhang et al. Citation2020). As shown in . kuehnei was positioned near T. sexlineatus, and had a closer relationship with four species within the genus Takydromus, including T. septentrionalis, T. wolteri, T. amurensis, and T. tachydromoides. Meanwhile, the phylogenetic relationship of T. kuehnei is separated from the genus Eremias, and far away from Phynocephalus putjatai which is not the species of family Lacertiadae.
Author contributions
L.X.W and G.H.D were involved in the conception and design, analysis and interpretation of the data. G.H.D collected the sample. L.X.W. and K.N.L. wrote the original paper. G.H.D. finally approved the version to be published.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MZ435950. The associated BioProject, BioSample, and SRA numbers are PRJNA636742, SAMN19899855, and SRR14933831, respectively.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kim IH, Park J, Cheon KS, Lee HJ, Kim JK, Park D. 2016. Complete mitochondrial genome of Schlegel’s Japanese gecko Gekko japonicus (Squamata: Gekkonidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3684–3686.
- Park J, Koo KS, Kim IH, Park D. 2016. Complete mitochondrial genomes of Scincella vandenburghi and S. huanrenensis (Squamata: Scincidae). Mitochondrial DNA B Resour. 1(1):237–238.
- Uetz P, Freed P, Aguilar R, Hošek J. 2021. The Reptile Database. [EB/OL]. http://www.reptile-database.org.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.