Abstract
Eopsetta grigorjewi (Pleuronectiformes: Pleuronectidae) is a demersal flatfish found in South Korea, Japan, Taiwan, China, and the Yellow Sea. E. grigorjewi complete mitochondrion DNA (mtDNA) consists of 16,921 bp and a 54% A + T content. It includes 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA), 13 protein-coding genes, and 1 non-coding regulatory area. ND2, ND3, ND4, COII, COIII, ATPase6, and CytB all have incomplete stop codon genes. The evolutionary analysis of 13 species from the same family indicated a close relationship. This work will be valuable for future research on molecular evolution and the creation of biomarker databases for determining the originality of E. grigorjewi.
Eopsetta grigorjewi (Herzenstein, 1890) is a member of the Pleuronectiformes order, Pleuronectidae family, and Pleuronectinae subfamily (Froese and Pauly Citation2012). It is also known as mushigarei, round nose flounder, shothole flounder, and shotted halibut (Robins et al. Citation1991). It resides near the sublittoral zone at depths of 200–4000 feet, has an adult fish length of around 24 inches, and female fishes are bigger than male fishes (Kim et al.Citation2011). These fish are East Asian marine fauna found in the Western Pacific Ocean, including the east coast of the Republic of Korea, the north coast of Japan, the south shore of Taiwan, China, and the Yellow Sea (Froese and Pauly Citation2018). Currently, the genus Eopsetta is known to include two species: E. grigorjewi and E. jordani Lokington, 1879. (Froese and Pauly Citation2012). Fish is high in protein, fatty acids, and minerals. The majority of Koreans consume seafood, and the eating culture is similar to that of Japan and China. People consume fish that has been cooked with seasonings, fried, or consumed raw. In the sea fish industry, flatfish are very important for export/import. It is critical to distinguish between imported and exported fish origins. In this study, we are focusing on characterizing mitochondrial DNA (mtDNA), which will aid in the development of molecular identification tools and will be a useful resource in the future to study genetic diversity and molecular evaluation.
The specimen with voucher specimen number MFDS-FGA12 was captured by a trawl net off the coast of Busan (East Sea), Republic of Korea (35°06′28.1″N 129°12′40.3″E). It was deposited at the Department of Food Engineering, Pukyong National University, Busan, Republic of Korea (Ji-Young Yang, [email protected]). Following the manufacturer's recommendations, genomic DNA was produced using the DNeasy Blood and Tissue Kit, Qiagen, Germany. The DNA library was produced and sequenced on the MGISEQ-2000 system utilizing paired-end reads (150 bp) using the MGIEasy DNA Library Prep Kit (MGI, China). Cutadapt ver. 1.9 (Martin Citation2011) was used to clean the raw reads, and the cleaned data was mapped using CLC Genomics Workbench (Qiagen, Germany). The final mtDNA sequence was annotated using the MitoFish-database of the fish mitochondrial genome (Sato et al. Citation2018). The MEGA11 platform was used to construct a Pleuronectidae phylogenetic relationship tree using the maximum likelihood method and the Tamura-Nei model with 1000 bootstraps (Tamura et al. Citation2021).
The whole genome was sequenced using 32,704,409 read-pairs, yielding 9,811,322,700 bp. The whole mtDNA of E. grigorjewi is close-circular, with 16,921 bp deposited in GenBank (Accession number OK545542.1). It has a 54% A + T content, and the individual base-pair composition includes A 27% (4635 bp), T 27% (4309 bp), G 17% (2915 bp), and C 29% (5062 bp). Two ribosomal RNA (rRNA), twenty-two transfer RNA (tRNA), thirteen protein-coding genes, and one regulatory area are among the 38 components of E. grigorjewi mtDNA. Nine of these genes, notably ND6 and eight tRNAs, were found on the L-strand (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro). At the present, mtDNA has been shown to contain incomplete genes, including the stopping codons of ND2, ND3, ND4, COII, COIII, Cyt B, and ATPase 6, and atypical codon usage in tRNA-Ser (GCU, UGA) and tRNA-Leu (UAG, UAA).
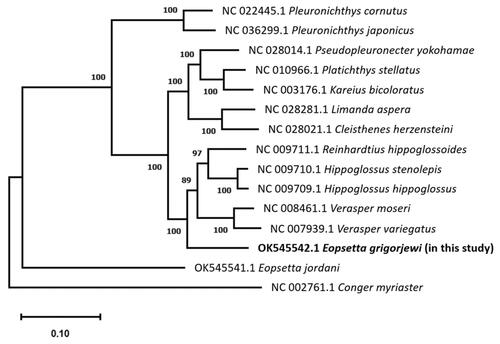
Phylogenetic relationships tree within the Pleuronectidae family were built using a published mtDNA dataset, as shown in . The phylogenetic study revealed that E. grigorjewi (Accession no. OK545542.1) is closely related to other genera and species in the Pleuronectidae family. For a better understanding of the species phylogeny, a detailed morphological and molecular phylogenetic investigation is required. These findings will help in understanding the genetic diversity and evolution of Eopsetta and Pleuronectidae. The present study's mtDNA results will be useful for future research on molecular evolution and the construction of a biomarkers database for determining the originality of E. grigorjewi.
Figure 1. A phylogenetic tree within family Pleuronectidae, constructed with mtDNA data from NCBI GenBank of 13 species, Eopsetta grigorjewi (present study) and Conger myriaster was set as the outgroup using MEGA11 (ClustalW, maximum likelihood method and Tamaru-Nei model) with 1000 bootstrap replicates. Bootstrap possibilities are indicated by numbers on each node.
Ethical approval
The sample used for this study was a dead body of fish and as per the animal experimental ethics in the Republic of Korea (Standard operating guideline; IACUC – Institutional Animal Care and Use Committee, Book no. 11-1543061-000457-01, effective from Dec. 2020) does not need any approval from Ethics Committee.
Author contributions
Conceptualization, J-O.K., and G-D.K.; Formal analysis and interpretation, M.P.P., and J-O.K.; Software, M.P.P., Y.B.S., and J.S.; Validation, J-O.K., and J-Y.Y.; Data curation, M.P.P., and J-O.K.; Writing-original draft, M.P.P., and J-O.K.; Writing-review and editing, J.S., Y.B.S., J-Y.Y., and G-D.K.; Supervision, J-Y.Y., and G-D.K.; Funding acquisition, G-D.K. All authors reviewed and agreed for publication.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OK545542.1. The associated BioProject, BioSample, and SRA numbers are PRJNA794315, SAMN24624474, and SRP353559, respectively.
Additional information
Funding
References
- Froese R, Pauly D, eds. 2012. Pleuronectidae. In FishBase. October 2012 version.
- Froese R, Pauly D, eds. 2018. Eopsetta grigorjewi. In FishBase. February 2018 version.
- Kim YH, Kim YS, Kang HJ, Kim JK, Chun YY. 2011. Age and growth of shotted halibut Eopsetta grigorjewi in the East China Sea. Korean J Ichthyology. 23(1):861–36 (in Korean with English abstract).
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17(1):10–12.
- Robins CR, Bailey RM, Bond CE, Brooker JR, Lachner EA. 1991. World fishes important to North Americans exclusive of species from the continental waters of the United States and Canada. Amer Fish Soc Spec Publ. 21:243.
- Sato Y, Miya M, Fukunaga T, Sado T, Iwasaki W. 2018. MitoFish and MiFish Pipeline: a mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol Biol Evol. 35(6):1553–1555.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
