Abstract
In this study, we sequenced the complete mitochondrial genome (mitogenome) of Cerogria popularis Borchmann, 1936 based on the Illumina platform. The circular DNA molecule is 16,175 bp in size, including the 37 typical animal mitochondrial genes and a non-coding control region. All 37 genes are arranged in the same order as the previously reported most mitogenomes of Tenebrionidae. All PCGs initiate with standard start codon ATN (ATA/T/G/C) except cox1 with the special start codon AAT. Most PCGs terminate with TAA/G, whereas cox1, atp6, nad5, and nad4 end with its incomplete form T-. All the 22 tRNAs have the typical clover-leaf structure except for trnS1. The rrnL and rrnS genes are 1,250 and 737 bp in length, with an AT content of 82.6 and 84.5%, respectively. The phylogenetic tree supports the monophyly of the included tenebrionid subfamilies represented by more than one species. Furthermore, the sister relationship between Lagriinae and other tenebrionid subfamilies is recovered.
Keywords:
The subfamily Lagriinae belongs to the family Tenebrionidae of Coleoptera, and to date there are more than 2,200 species (subspecies) described worldwide (Zhou and Chen Citation2014). Its range and taxonomic status have been debated throughout the history of research (Zhu Citation2003). This study firstly sequenced the complete mitochondrial genome (mitogenome) of Cerogria popularis Borchmann, 1936, which will be useful for exploring the phylogenetic status of Lagriinae.
The samples of C. popularis were collected from Weining County (E104°36′39.63″, N26°48′44.16″), Guizhou Province, China and deposited in Insect Herbarium of Guizhou Academy of Forestry, Guiyang (GZAF-2021-CT0078) (URL, Kai Hu and [email protected]). Identification of adult specimens was based on morphological characteristics (Zhu Citation2003; Zhou Citation2011). The total genome DNA was sequenced using Illumina MiSeq format (Illumina, San Diego, CA). Then the data was assembled by NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) with the cox1 gene of Chrysomela vigintipunctata (NC_050933) as the seed. The complete mitogenome was annotated using MITOZ v1.04 (Meng et al. Citation2019). All 13 protein-coding gene sequences were aligned using MAFFT v7.394 (Kuraku et al. Citation2013) with L-INS-i (accurate) strategy. Maximum likelihood (ML) tree was inferred by IQ-TREE v1.6.3 (Nguyen et al. Citation2015) under the optimal model (GTR + I + G for Subset1 (cox2, cytb, cox3, nad3, and atp6), Subset2 (nad2, nad6, and atp8), Subset3 (cox1), and Subset4 (nad1, nad4L, nad4, and nad5)).
The mitogenome of C. popularis (GenBank accession no. MZ699994) is 16,175 bp in size, which comprises 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and a non-coding control region (length: 1,692 bp). All 37 genes are arranged in the same order as the previously sequenced most mitogenomes of Tenebrionidae (Hong et al. Citation2020). The overall nucleotide composition of the newly sequenced mitogenome is 41.9% T, 10.5% C, 38.3% A, and 9.3% G, with an AT content of 80.2%. All PCGs initiate with standard start codon ATN (ATA/T/G/C) except cox1 with the special start codon AAT. Most PCGs terminate with TAA/G, whereas cox1, atp6, nad5, and nad4 end with its incomplete form T-. All tRNAs exhibit a typical clover-leaf secondary structure except for trnS1, in which the dihydrouridine (DHU) arm is replaced by a simple loop. The rrnL and rrnS genes are 1,250 and 737 bp in length, with an AT content of 82.6 and 84.5%, respectively. The control region is located between rrnS and trnI, which is 1,692 bp in length.
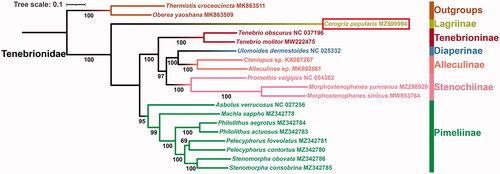
In this study, based on the nucleotide data of 13 PCGs from 17 Tenebrionidae species and two outgroup taxa from Cerambycidae, we reconstructed the ML phylogenetic tree (). The phylogenetic analyses strongly support the monophyly of the included tenebrionid subfamilies represented by more than one species (Alleculinae, Stenochiinae, Tenebrioninae, and Pimeliinae) (BS ≥95), similar to the recent study (Hong et al. Citation2020), but our taxon sample is much larger. In Tenebrionidae, the relationships among included subfamilies are inferred as (Lagriinae + (Pimeliinae + (Tenebrioninae + (Stenochiinae + (Alleculinae + Diaperinae))))). Furthermore, the sister relationship between Lagriinae and other tenebrionid subfamilies is recovered (BS = 100).
Ethical approval
Experiments were performed following the recommendations of the Ethics Committee of Guizhou Academy of Forestry. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
Conceived and designed the experiments: Kai Hu and Jiansheng Qiu. Performed the experiments: Tongtong Liu and Yuekai Wu. Analyzed the data: Kai Hu. Wrote the paper: Tongtong Liu and Jiansheng Qiu. Helped to proofread the paper: Kai Hu and Yuekai Wu. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ699994. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA763012, SRR15890679, and SAMN21419663, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hong KJ, Ki W, Lee HB, Park JS, Lee WH. 2020. The second complete mitochondrial genome of Alphitobius diaperinus Panzer, 1797 (Coleoptera: Tenebrionidae): investigation of intraspecific variations on mitochondrial genome. Mitochondrial DNA B Resour. 5(3):794–2981.
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41(Web Server issue):W22–W28.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Zhou Y, Chen B. 2014. Five new record species in Lagriinae from Chongqing. Journal of Chongqing Normal University. 31(6):29–33.
- Zhou Y. 2011. Taxonomy of Lagriina from China (Coleoptera, Tenebrionidae, Lagriini) [master’s thesis]. Shijiazhuang: Hebei University; p. 1–174.
- Zhu YX. 2003. Morphological and taxonomic study on the subfamily Lagriinae of China (Coleoptera: Lagriidae) [master’s thesis]. Chongqing: Southwest Agricultural University; p. 1–129.