Abstract
Magnolia coriacea, Chang et B. L. (Magnoliaceae) is a critically endangered tree, endemic to Yunnan province, China. In this study, the complete chloroplast genome of M. coriacea was sequenced and analyzed. The total chloroplast genome size of M. coriacea is 160,113 bp, including a pair of inverted repeat regions (IRs, 26,576 bp) separated by a large single-copy (LSC, 88,175 bp) region and a small single-copy region (SSC, 18,786 bp). The complete chloroplast genome contains 86 protein-coding (PCGs), 37 transfer RNA (tRNAs), and 8 ribosomal RNA (rRNAs) genes. The phylogenetic analysis showed that M. coriacea is closely related to M. cathcartii. This study contributes to the bioinformatics on the evolution, genetic, conservation, and molecular biology for future studies of Magnoliaceae.
Magnolia is a genus of the family Magnoliaceae, comprising over 60 species; mainly distributed in tropical, subtropical, and temperate regions of Asia (Zhao et al. Citation2009). There are 32 species currently recognized in China, mainly distributed in the southwest and southeast regions (Cicuzza et al. Citation2007). Yunnan Province harbors a high number of Magnoliaceae species, many of which are at risk of extinction due to habitat destruction (Fu and Jin Citation1992; Cicuzza et al. Citation2007). Among these species, one of the most threatened is M. coriacea, Chang et B. L. Chen Citation1988, which is endangered by habitat destruction, fragmentation, and degradation while its distribution range is entirely unprotected (Tang et al. Citation2011). M. coriacea was described recently and is endemic to Southeast Yunnan, China (Chen Citation1988; Liu and Wu Citation1988). This species was said to be similar to M. martinii and bears scented whitish or creamy yellow flowers with 6–7 tepals and anthesis lasts from February to April (Chen Citation1988; Sun and Yan Citation2006; Zhao and Sun Citation2009). The existing population of this species is restricted to limestone outcrops in just a few localities at 1300–1700 m (Cicuzza et al Citation2007; Zhao and Sun Citation2009; Zhao et al. Citation2009; Tang et al. Citation2011). Most of the individuals in the extant populations normally bear flowers, with estimated fruit set and fertile seed production rates at only 6.7% and 0.2%, respectively (Zhao and Sun Citation2009). M. coriacea has been evaluated as a critically endangered tree based on IUCN criteria (Cicuzza et al. Citation2007). At present, there are no published DNA sequences of M. coriacea in GenBank. Here, we assembled and analyzed the complete chloroplast genome of M. coriacea using high-throughput sequencing methods. This study contributes to the bioinformatics on the evolution, genetic, conservation, and molecular biology for future studies of Magnoliaceae ().
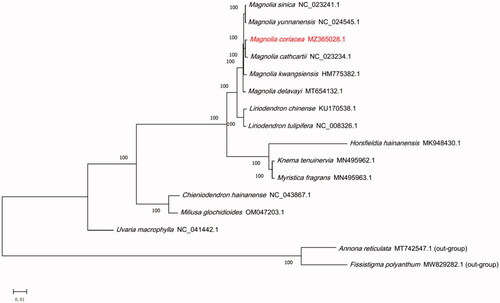
Figure 1. The maximum-likelihood (ML) phylogenetic tree for M. coriacea based on 16 chloroplast genome sequences of Magnoliales. Numbers on the nodes are bootstrap values from 1000 replicates.
Fresh young leaves of M. coriacea were obtained from Malipo County, Wenshan City, Yunnan Province, China (104° 51′ 14″ E, 23° 10′ 3″ N, 1480 m altitude) and DNA samples were stored at Guizhou Normal University (contact person and email: Baizhu Li, [email protected]; accession number: HX-001). Total genomic DNA was extracted using the CTAB method (Li et al. Citation2013). DNA was sent to Shanghai Biozeron Co., Ltd. (Shanghai, China) to construct the DNA library and sequencing using the Illumina HiSeq 6000 sequencing platform. The low quality sequences were removed using Trimmomatic0.39 using the default settings (Bolger et al. Citation2014). The trimmed reads were selected for the cp genome assembly with the NOVOPlasty4.2 software using the default setting (Dierckxsens et al. Citation2016). The complete chloroplast genome of M. coriacea was annotated by GeSeq (Tillich et al. Citation2017). The complete cp genome was submitted to GenBank (https://www.ncbi.nlm.nih.gov/search/all/?term=MZ365028; accession number of MZ365028).
The complete cp genome sequence of M. coriacea is 160,113 bp and shows a characteristic quadripartite circular structure. The GC content of the complete genome is 39.24%. The large single-copy (LSC) (GC, 37.92%) region is 88,175 bp and the small single-copy (SSC) (GC, 34.26%) region is 18,786 bp. These are separated by a pair of IRa and IRb regions of 26,576 bp. The chloroplast genome of M. coriacea has 131 genes in total, including 86 protein-coding, 37 transfer RNA (tRNA), and eight ribosomal RNA (rRNA) genes. Among these, duplication events occurred with 17 genes (ndhB, rpl2, rpl23, rps7, rps12, ycf1, ycf2, trnM-CAU, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, rrn16, rrn23, rrn4.5, and rrn5). In addition, 246 simple sequence repeat loci were identified in the cp genome. These were mainly composed of mononucleotide (A, 19.1%; T,26.8%) and trinucleotide (29.67%) repeats by MISA-web (Thiel et al. Citation2003; Beier et al. Citation2017). We compared the basic characteristics of the complete chloroplast genome of the genus M. coriacea with those of 17 other currently published species of the genus Magnolia, and found that they all have a characteristic quadripartite circular structure, the size (160,113 bp) and GC content (39.24%) of M. coriacea were not significantly different from those of the genus Magnolia species (Hall Citation1999; Li Citation2021).
In order to further determine the phylogenetic position of M. coriacea, 16 cp genome sequences from Magnoliales family were downloaded from GenBank, and two Annonaceae family species were selected as outgroup. Based on MAFFT function under BioAider1.0 software was used for the alignment of cp genome sequences (Zhou et al. Citation2020). A maximum-likelihood (ML) tree was constructed using PhyML 3.0 online website with the GTR model and 1000 bootstrap replicates (Guindon et al. Citation2010). The phylogenetic analysis showed that M. coriacea was closely related with M. cathcartii, and they form a group with other seven species of the genus Magnoliaceae. These data provide important bioinformatic resources for species identification, and phylogenetic relationships within the Magnoliales.
Ethical approval
The authors comply with the International Union for Conservation of Nature (IUCN) policies research involving species at risk of extinction, the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. Our study has been approved by local authorities (Malipo County Endangered Wild Medicinal Plant protection and Breeding Association). Xibin Guo is the Organization manager (e-mail address: [email protected]).
Authors contributions
Conceptualization: Yu Li and Baizhu Li; methodology: Yu Li, Baizhu Li, Zhaofeng Li, Yi Yin, and Xiaoxin Tang; resources: Yu Li, Baizhu Li, Zhaofeng Li, Yin Yi, and Xiaoxin Tang; writing-original draft preparation: Baizhu Li, Yu Li, and Xiaoxin Tang; writing-review and editing: Baizhu Li, Yu Li, and Xiaoxin Tang; supervision: Baizhu Li, Yu Li, and Xiaoxin Tang; funding acquisition: Yin Yi and Xiaoxin Tang; all authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/search/all/?term=MZ365028 under the accession no. MZ365028. The associated BioProject, SRA, and BioSample numbers are PRJNA774168, SRR16563234, and SAMN22556078, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):801–2120.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Chen BL. 1988. Four new species of Michelia from Yunnan. Acta Sci Nat Univ Sunyatseni. 3:86–91.
- Cicuzza D, Newton A, Oldfield S. 2007. The Red List of Magnoliaceae. Cambridge (UK): Fauna & Flora International; p. 20.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 4:gkw955.
- Fu LK, Jin JM. 1992. China plant red data book-rare and endangered plants. Beijing, China: Science Press.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Liu YH, Wu RF. 1988. Some new taxa of Michelia from China. Acta Bot Yunnanica. 10:335–342.
- Li JL, Wang S, Yu J, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48(1):72–78.
- Li YY, Zhou M, Wang LM, Wang JQ. 2021. The characteristics of the chloroplast genome of the Michelia chartacea (Magnoliaceae). Mitochondrial DNA B Resour. 6(2):493–495.
- Sun WB, Yan L. 2006. Unpublished report on global trees campaign fieldwork in China. Cambridge (UK): FFI.
- Tang CQ, He LY, Gao Z, Zhao XF, Sun WB, Ohsawa M. 2011. Habitat fragmentation, degradation, and population status of endangered Michelia coriacea in Southeastern Yunnan, China. Mt Res Dev. 31(4):343–350.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht JES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W1.
- Thiel T, Michalek W, Varshney R, Graner A. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 106(3):411–422.
- Zhao X, Sun W. 2009. Abnormalities in sexual development and pollinator limitation in Michelia coriacea (Magnoliaceae), a critically endangered endemic to Southeast Yunnan, China. Flora. 204(6):463–470.
- Zhao XF, Sun WB, Yang JB, Jing M. 2009. Isolation and characterization of 12 microsatellite loci for Michelia coriacea (Magnoliaceae), a critically endangered endemic to Southeast Yunnan. Conserv Genet. 10(5):1583–1585.
- Zhou ZJ, Qiu Y, Pu Y, Huang X, Ge XY. 2020. BioAider: an efficient tool for viral genome analysis and its application in tracing SARS-CoV-2 transmission. Sustain Cities Soc. 63:102466.
