Abstract
The complete chloroplast genome sequence of Clematis mandshurica Ruprecht (1867), a specie of the Ranunculaceae family, and its phylogenetic relationships with other species have been reported in this study. The complete chloroplast genome of C. mandshurica is 159,563 bp in length, including a large single-copy (LSC) region of 79,360 bp, a small single-copy (SSC) region of 18,121 bp, and a pair of identical inverted repeat regions (IRs) of 31,041 bp. The genome encodes a total of 132 genes, including 90 protein-coding genes, 34 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The phylogenetic analysis reveals that C. mandshurica was found to be closest to Clematis taeguensis. The complete chloroplast genome of C. mandshurica contributes to a better understanding of phylogenetic relationships among Clematis species.
Clematis is a cosmopolitan genus of about 355 species in the Ranunculaceae (Wang and Li Citation2005; Yang et al. Citation2019; Chen et al. Citation2021), which is widely distributed throughout the world. Clematis mandshurica root is the most famous traditional Chinese herbal medicines obtained from Clematis species (Hao et al. Citation2013), widely used as Wei Ling Xian in traditional medicine (Lee et al. Citation2014) and is native to China, Korea, Mongolia, and the Russian Far East (Dong et al. Citation2016). The dried roots and rhizomes of C. mandshurica have been used as an analgesic, anti-inflammatory, anti-tumor, and dispel wind agent (Huang Citation1999; He et al. Citation2011; Ling et al. Citation2013; National Pharmacopoeia Committee Citation2020). At present, the research on C. mandshurica mainly focused on cultivation techniques (Han et al. Citation2011, Citation2021), chemical constituents and pharmacological activities (Lee et al. Citation2020; Lin et al. Citation2021). However, the genetics and molecular biology of C. mandshurica are poorly understood. Therefore, we here report the first complete chloroplast genome of C. mandshurica by high throughput sequencing technology, which will provide valuable bioinformatic data for understanding the systematics of C. mandshurica and genetic research.
Fresh leaves of C. mandshurica were provided from Medicinal Herb Garden, Jilin Agricultural University, Changchun, China (43°48′23″N, 125°24′57″E). The voucher specimen was deposited in the Herbarium of College of Chinese Medicinal Materials, Jilin Agricultural University (https://zhongyao.jlau.edu.cn; Zeliang Lü, [email protected]) under the voucher number Y. Cui 2021009. Genomic DNA was extracted by a QIAquick Gel Extraction kit (Qiagen, Hilden, Germany). Pair-end raw reads were obtained by PE 150 library and the Illumina Hiseq 2500 platform. Finally, the raw data (1.1 Gb) were obtained. Genome assembly and annotation were conducted using metaSPAdes9 (Nurk et al. Citation2017) and CPGAVAS2 (Shi et al. Citation2019), respectively. The annotated cp genome sequence was submitted to GenBank under the accession number OK375873.
The length of complete chloroplast genome of C. mandshurica was 159,563 bp, displaying a large single-copy (LSC), a small single-copy (SSC), and a pair of inverted repeat (IR) regions of 79,360 bp, 18,121 bp, and 31,041 bp, respectively. A total of 132 genes were annotated, including 90 protein-coding genes, 34 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The GC content of the cp genome was 37.98%, and the GC content of the LSC, SSC, and IR regions were 36.3%, 31.3%, and 42.1%, respectively.
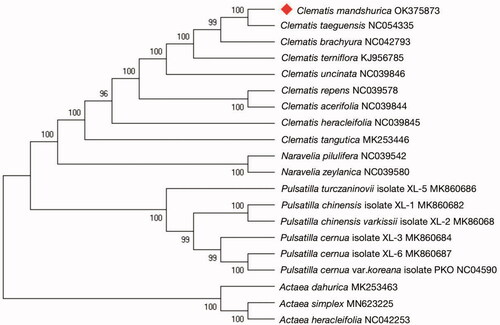
The maximum-likelihood (ML) phylogenetic tree was generated based on the complete genome of C. mandshurica and other species of the Ranunculaceae (). The 20 complete chloroplast sequences were aligned using MAFFT software (Katoh and Standley Citation2013), and a phylogenetic tree was constructed using MEGAX (Kumar et al. Citation2018) with a generalized time-reversible (GTR) sequence evolution model and an ML for tree improvement. Clematis mandshurica was found to be closest to Clematis taeguensis in tribe Anemoneae, subfamily Ranunculoideae. The phylogenetic analysis resolved great chloroplast divergence within the genus Clematis (Kyun et al. Citation2021). In conclusion, the complete chloroplast genome of C. mandshurica contributes to a better understanding of the phylogenetic relationships among Clematis species, which can contribute important information for understanding phylogenetic and evolutionary studies of Ranunculaceae.
Ethical approval
The materials used in this study are not included in IUCN red list, the collection area is not a protected area. Research and collection of plant material was conducted according to the guidelines provided by JLAU (Jilin Agricultural University). Permission was granted by the China Biotechnology Center and Science and Technology Department of Jilin Province.
Author contributions
Zhongming Han, Limin Yang, and Yunhe Wang: conceptualized and designed research; Yi Cui, Lihua Yang, and Mei Han: analyzed data and wrote original draft of the manuscript; Yanzhe Ding, Yingxin Sun, Jiao Wang, and Yunfei Xi: contributed to research materials and to the draft manuscript. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OK375873. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA764578, SRR16071938, and SAMN21841556, respectively.
Additional information
Funding
References
- Chen X, Chang Q, Xia P, Liang Z, Yan K. 2021. The complete chloroplast genome of Clematis henryi var. ternata (Ranunculaceae). Mitochondrial DNA B Resour. 6(4):822–1320.
- Dong Y, Zhu L, Bai Y, Ou Y, Wang C. 2016. Two new eriophyid mite species associated with Clematis terniflora var. mandshurica in China (Acari, Eriophyidae). ZooKeys. 637(621):1–14.
- Han Z, Tian X, Zhang F, Wang Y, Han M, Yang L. 2021. Effects of shading on growth, physiological characteristics and quality of Clematis manshurica. J Jilin Agric Univ. 43(3):303–309.
- Han Z, Zhao S, Liu C, Wang Y, Han M, Yang L. 2011. Effects of shading on growth and quality of triennial Clematis manshurica Rupr. Acta Ecol Sin. 31(20):6005–6012.
- Hao D, Gu X, Xiao P, Peng Y. 2013. Chemical and biological research of Clematis medicinal resources. Chin Sci Bull. 58(10):1120–1129.
- He Y, Li L, Zhang K, Liu Z. 2011. Cytotoxic triterpene saponins from Clematis mandshurica. J Asian Nat Prod Res. 13(12):1104–1109.
- Huang KC. 1999. The pharmacology of Chinese herbs. Louisville: CRC Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kyun PB, Ghimire B, Dong CS. 2021. Taxonomic identity of Korean endemic Clematis taeguensis Y. N. Lee (Ranunculales: Ranunculaceae). J Asia-Pac Biodivers. 14(1):70–77.
- Lee CW, Park SM, Kim YS, Jegal KH, Lee JR, Cho IJ, Ku SK, Lee JY, Ahn Y, Son Y, et al. 2014. Biomolecular evidence of anti-inflammatory effects by Clematis mandshurica Ruprecht root extract in rodent cells. J Ethnopharmacol. 155(2):1141–1155.
- Lee T, Park JH, Kim B, Park YE, Lee J, Ahn JH, Park CW, Noh Y, Lee J, Kim S, et al. 2020. YES-10, a combination of extracts from Clematis mandshurica RUPR. and Erigeron annuus (L.) PERS., prevents ischemic brain injury in a gerbil model of transient forebrain ischemia. Plants. 9(2):154.
- Lin T, Wang L, Zhang Y, Zhang J, Zhou D, Fang F, Liu L, Liu B, Jiang Y. 2021. Uses, chemical compositions, pharmacological activities and toxicology of Clematidis Radix et Rhizome – a review. J Ethnopharmacol. 270:113831.
- Ling L, Mei-Ling G, Yu-Xin H. 2013. Mandshunosides C–E from the roots and rhizomes of Clematis mandshurica. Phytochem Lett. 6(4):570–574.
- National Pharmacopoeia Committee. 2020. Pharmacopoeia of the People's Republic of China. Vol. I. Beijing: China Medical Science Press.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Wang WT, Li LQ. 2005. A new system of classification of the genus Clematis (Ranunculaceae). Acta Phytotaxon Sin. 43(2):115–121.
- Yang Y, Guo X, Wang K, Liu Q, Liu Q. 2019. Anther and ovule development in Clematis terniflora var. mandshurica (Ranunculaceae). Flora. 253:67–75.