Abstract
Ranunculus pekinensis (L. Liou) Luferov 1997, a perennial aquatic herb, is endemic to Beijing, China and has high water quality requirements. Because its habitat is under great threat and its population is declining, it is now listed as a national protected plant in China. To provide genomic resources for future research of this endangered species, the complete chloroplast genome sequence of R. pekinensis was assembled and annotated for the first time. The complete chloroplast genome sequence was 156,139 bp in length, containing a large single copy region (LSC) of 85,430 bp and a small single copy region (SSC) of 19,970 bp, which were separated by a pair of 25,367 bp inverted repeat regions (IRs). The complete chloroplast sequence contained 112 unique genes, including 30 tRNA, 4 rRNA, and 78 protein-coding genes. The overall guanine-cytosine (GC) content of the chloroplast genome was 37.8%, and the GC contents of the LSC, SSC, and IR regions were 36.0%, 31.3%, and 43.5%, respectively. Phylogenetic analysis with the reported chloroplast sequences showed that R. pekinensis was closely related to R. bungei Steud. 1841, both of which belonged to Ranunculus Sect. Batrachium DC. 1817. These data will provide essential resources regarding the evolution and conservation of R. pekinensis.
Ranunculus L. 1753 (Ranunculaceae) is widely distributed on all continents except for Antarctica, mainly in north temperate regions, with about 550 species in total (Wang and Michael Citation2001). Ranunculus pekinensis (L. Liou) Luferov 1997, a perennial aquatic herb, is endemic to Beijing, China, and grows in valley streams at altitudes of 80–1200 m (Wu et al. Citation2014). This species belongs to Ranunculus Sect. Batrachium DC. 1817 and has high water quality requirements (Han et al. Citation2020). Over the past 30 years, the continuous drought in this area has left many streams depleted of water, which has led directly to habitat loss. Water pollution caused by domestic waste and herbicides also seriously threatens the survival and reproduction of R. pekinensis (Wu et al. Citation2014). Recently, R. pekinensis was listed as a national protected plant species. Therefore, it is vital to implement effective science-based protection measures to expand the population size.
Chloroplast in plant cells is a double membrane-bounded organelle that plays vital metabolic roles including photosynthesis, amino acid and lipid synthesis. In most plant species, the chloroplasts (cp) have a circular genome, varying in size from 72 to 217 kb and containing about 130 genes. And the chloroplast genome has a quadripartite structure in which a pair of inverted repeats (IRs) region separates a large single copy (LSC) from small single copy (SSC) regions in most species. Numerous mutational events take place in chloroplast genomes, including substitutions, insertions and deletions (InDels), inversions, genome rearrangements, and translocations. Polymorphism in chloroplast genomes has been exploited to resolve taxonomic and phylogenetic discrepancies. Moreover, it is also useful for development of DNA barcodes, population genetics and evolutionary studies (Mehmood et al. Citation2020). However, to date, no chloroplast genome resources have been available for R. pekinensis. In this study, the complete chloroplast genome of R. pekinensis was assembled to provide a genetic foundation for further research.
Fresh young leaves of R. pekinensis were collected from Yudu Mountain in Beijing, China (N 40°33′7.38″, E 115°52′43.73″), the specimens were deposited at the Herbarium of Beijing Forestry University (http://bjfc.bjfu.edu.cn, Liangcheng Zhao: [email protected]) under voucher number LXH001. The CTAB method (Doyle and Doyle Citation1987) was used to extract the total genome DNA. Then, an Illumina HiSeq 4000 platform at Novogene (http://www.novogene.com, China) was used to perform 2 × 150 bp pair-end sequencing. The Map to Reference function in Geneious Prime (Kearse et al. Citation2012) was used to exclude nuclear and mitochondrial reads using the published plastid genome of R. bungei Steud. 1841 as a reference (Accession no. MK253468). The filtered chloroplast reads were then de novo assembled (with ‘Low Sensitivity/Fastest’ sensitivity) and concatenated into larger contigs using the Repeat Finder function in Geneious Prime. The raw reads were again mapped to the larger contigs (with the Map to Reference function) to extend their boundaries until all contigs could be concatenated into one contig. Gaps were bridged using the Fine Tuning function of Geneious Prime. The IR region was determined using the Repeat Finder function in Geneious Prime. The mean coverage depth of the assembly was 765×. The assembled chloroplast sequence was annotated using the Plastid Genome Annotator (PGA, Qu et al. Citation2019) and verified by Geneious Prime. The final R. pekinensis chloroplast genome sequence was then submitted to GenBank under the accession number of OK166810.
The chloroplast genome of R. pekinensis was 156,139 bp in length, containing a large single copy region (LSC) of 85,430 bp and a small single copy region (SSC) of 19,970 bp, which were separated by a pair of 25,367 bp inverted repeat regions (IRs). The chloroplast genome sequence contained 112 unique genes including 30 tRNA genes, 4 rRNA genes, and 78 protein-coding genes. The overall guanine-cytosine (GC) content of the chloroplast genome was 37.8%, and the GC contents of the LSC, SSC, and IR regions were 36.0%, 31.3%, and 43.5%, respectively.
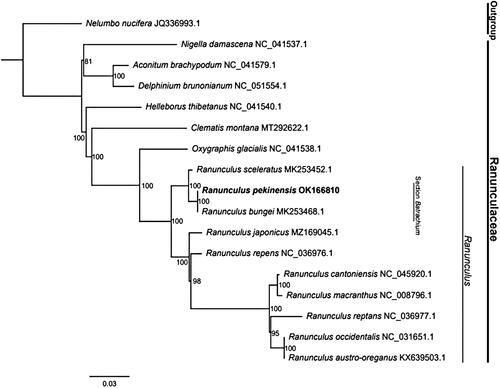
To understand the phylogenetic relationship between R. pekinensis and its related taxa, the complete chloroplast genome sequences of nine Ranunculus species and seven other species in Ranunculaceae were downloaded from the NCBI GenBank. All sequences were aligned using MAFFT (Katoh et al. Citation2017). The maximum likelihood (ML) method was used for phylogeny reconstruction. The sequence alignment was performed as described by Liu et al. (Citation2018). Then, the ML tree was constructed using IQ-TREE software (Kalyaanamoorthy et al. Citation2017; Hoang et al. Citation2018). The phylogenetic analysis showed that all Ranunculus species clustered together and were closely related to Oxygraphis Bunge 1835. They all belonged to the Trib. Ranunculeae of Ranunculaceae. In addition, R. pekinensis and R. bungei were united with strong support (ML BP = 100), and were robustly nested in the clade of Ranunculus (). This result was consistent with previous studies (Khatere et al. Citation2010; Gerhard and Alexander Citation2017), supporting the conclusion that Batrachium (DC.) Gray 1821 should be treated as a section of the genus Ranunculus. These data will provide essential resources regarding the evolution and conservation of R. pekinensis.
Ethics statement
This work was part of the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, China. Therefore, field studies, including the collection of plant material have been carried out in accordance with local permissions and national regulations.
Author contributions
Xuehua Liu, Wenli Yang, and Gangmin Zhang contributed to design of the study and field investigation; Xuehua Liu, Hao Liu and Yueming Cui performed the experiment; Xuehua Liu, Danke Zhang and Lei Wang contributed to the analysis and interpretation of the data; Xuehua Liu, Hongyu Hu and Yue Yin contributed to the drafting of the paper; Gangmin Zhang contributed to revising the paper critically for intellectual content and the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Acknowledgement
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OK166810. The associated BioProject, SRA, and BioSample numbers are PRJNA770464, SRS10552956, and SAMN22218715 respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:841–15.
- Gerhard W, Alexander AB. 2017. A taxonomic account of Ranunculus section Batrachium (Ranunculaceae). Phytotaxa. 319(1):1–55.
- Han CH, Fan YQ, Jiang J, Song Z, Zhao XY. 2020. A study on the relationship between Batrachium pekinense L. Liou distribution and environmental features. Forest Resour Manag. 15(04):102–108.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Bioinformatics. 20(4):1160–1166.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Khatere E, Carlos L, Peter L. 2010. A molecular phylogeny, morphology and classification of genera of Ranunculeae (Ranunculaceae). Taxon. 59(3):809–828.
- Liu HJ, He J, Ding CH, Lyu RD, Pei LY, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Plant Sci. 9:1097.
- Mehmood F, Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2):1522–1530.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Wang WC, Michael GG. 2001. Ranunculus Linnaeus. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 6. Beijing: Science Press; p. 391–431.
- Wu JG, Jiang WJ, Fan YQ, Mu XY. 2014. Study on the hydro-environmental factors of the endangered species Batrachium pekinense L. Liou. Hubei for Sci Tech. 43(06):5–8.