Abstract
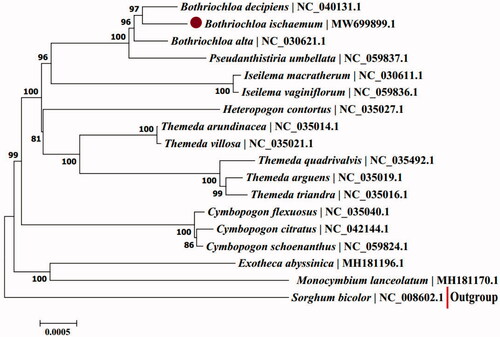
Bothriochloa ischaemum (Linn.) 1936 is a high-quality perennial forage in Loess Plateau of China. In this study, we sequenced and characterized the complete chloroplast genome of B. ischaemum, which was a circular DNA of 138,316 bp in length, including a large single copy (LSC) region of 80,226 bp, a small single copy (SSC) region of 12,526 bp, and the circular DNA was separated by a pair of identical inverted repeat regions (IRs) of 22,782 bp each. A total of 134 genes were identified, including 87 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Phylogenetic tree showed that B. ischaemum was closer to B. decipiens and B. alta, genus Bothriochloa was closely related to genus Pseudanthistiria. Our findings will be helpful for better understanding of genetic diversity of Bothriochloa plants.
Bothriochloa ischaemum (Linn.) 1936 is a high-quality perennial forage in Gramineae. It is the main building species of warm shrub grasslands in Loess Plateau of China. Bothriochloa ischaemum is characterized by soil fixation and water retention, strong vitality, drought resistance, high yield and grazing resistance (Li et al. Citation2019). However, the classification of B. ischaemum and its related species has always been controversial (Harlan and Wet Citation1963; Abdi et al. Citation2019). Complete chloroplast genome will provide the foundation for the species identification, germplasm diversity, genetic engineering of B. ischaemum. Our study aimed to establish and characterize the complete chloroplast (cp) genome of B. ischaemum, and provide additional effective data for phylogenetic study of Anthistiriinae in the future.
In this study, fresh leaves of B. ischaemum cv. ‘Taihang’ were sampled from experimental plots in Shanxi Agricultural University of China (112.5873° E, 37.4176° N). No specific permits were required for the collection of specimens for this study. ‘Taihang’ is the first cultivar of B. ischaemum which was registered by Chinese National Grass Variety Examination and Approval Committee in 2019, with the registration number of 568. ‘Taihang’ has large number of leaves, good quality and high grass yield, and which is more suitable for planting in northern and central China. The specimen was deposited at the herbarium of Shanxi Agricultural University (http://cyxy.sxau.edu.cn/, Lian-fen Li, [email protected]) under the voucher number SXAU-CoGS-19Bi01. Chloroplasts were isolated from 10-g fresh leaves by gradient centrifugation on Percoll. And then, chloroplast genomic DNA was extracted using CTAB method (Doyle and Doyle Citation1987). A 350 bp DNA library was constructed and then 150 bp paired-end reads were sequenced on an Illumina Hiseq 2500 platform. Approximately 3.3 Gb of clean bases were generated and assembled using NOVOPlasty v3.8.3 (Dierckxsens et al. Citation2017), using the psbB gene sequence of B. ischaemum as the seed sequence. Genome annotation was conducted using online tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017) and corrected manually.
The complete chloroplast genome of B. ischaemum (GenBank accession number: MW699899) had a circular DNA of 138,316 bp with GC content of 38.53%. And the circular DNA had a large single copy (LSC) region of 80,226 bp, a small single copy (SSC) region of 12,526 bp, and was separated by a pair of identical inverted repeat regions (IRs) of 22,782 bp each. In addition, 135 genes were annotated, including 87 protein-coding genes, 39 tRNA genes, and eight rRNA genes.
In order to reveal the phylogenetic position of B. ischaemum, a maximum-likelihood (ML) phylogenetic tree was constructed using 17 complete chloroplast genome sequences from Anthistiriinae Presl and one outgroup species (Sorghum bicolor). All of the 18 genome sequences were download from GenBank under corresponding accessions and were aligned using MAFFT v7.311 (Katoh and Standley Citation2013). The ML phylogenetic tree was produced by MEGA7 v7.0.26 (Kumar et al. Citation2016) using 1000 bootstrap replicates and the GTR + Gamma nucleotide substitution model. The phylogenetic tree showed that B. ischaemum was closer to B. decipiens and B. alta (). Analyzing the chloroplast genome, the genus Bothriochloa was closely related to the genus Pseudanthistiria in the Anthistiriinae Presl, this was the same as the results obtained by 2464 single-copy nuclear genes (Arthan et al. Citation2021). Analysis of the complete chloroplast genome of B. ischaemum is useful for studying the genetic diversity of Bothriochloa plants.
Authors’ contributions
YYL corrected genome annotation, wrote the draft of the manuscript, and was involved in writing of the final version of the manuscript. HYX performed data analysis and prepared the figure. HZ and ZQY collected samples of B. ischaemum, extracted DNA for next-generation sequencing, assembled and validated the genome. KHD conceived the study. FSX supervised all stages of writing of the manuscript. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number MW699899.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA718686, SRS8774889, and SAMN18559019 respectively.
Additional information
Funding
References
- Abdi S, Dwivedi A, Shashi Kumar S, Bhat V. 2019. Development of EST-SSR markers in Cenchrus ciliaris and their applicability in studying the genetic diversity and cross-species transferability. J Genet. 98:844.
- Arthan W, Dunning L, Besnard G, Manzi S, Kellogg E, Hackel J, Lehmann CER, Mitchley J, Vorontsova M. 2021. Complex evolutionary history of two ecologically significant grass genera, Themeda and Heteropogon (Poaceae: Panicoideae: Andropogoneae). Bot J Linn Soc. 196(4):437–455.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Harlan JR, Wet J. 1963. Role of apomixis in the evolution of the Bothriochloa-Dichanthium Complex1. Crop Sci. 3(4):314–316.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li CY, Dong J, Zhang XB, Zhong H, Jia HL, Fang ZH, Dong KH. 2019. Gene expression profiling of Bothriochloa ischaemum leaves and roots under drought stress. Gene. 691:77–86.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.