Abstract
Piper sarmentosum Roxb. (Piperaceae) is a traditional medicinal herb native to Southeast Asia. The complete genome of P. sarmentosum was sequenced and characterized in this study with the aim of providing genomic resources for the evolution and molecular breeding of P. sarmentosum. It has a typical quadripartite structure, with a large single-copy (LSC) region of 88,979 bp, a small single-copy (SSC) region of 18,274 bp, and two copies of 27,068 bp inverted-repeat regions (IRa and IRb). A total of 130 genes were annotated, comprising 85 protein-coding genes (PCGs), 8 ribosomal RNA (rRNA) genes, and 37 transfer RNA (tRNA) genes. The phylogenetic tree showed that P. sarmentosum in the current study is closely related to Piper longum.
Piper sarmentosum Roxb. (Piperaceae) is a traditional medicinal herb found mainly in Southeast Asia (Mathew et al. Citation2004). Due to its antioxidant and anti-inflammatory properties, it is beneficial for various diseases, including fever, toothache, dysentery, and traumatic injury (Sun et al. Citation2020). Its medicinal value is complemented by its edible value, as its leaves are frequently consumed as a tasty vegetable. Previous studies have demonstrated that the extracts of Piper sarmentosum are rich in alkaloids, essential oils, flavonoids, lignans, and steroids. The majority of current research on this plant has focused on compound isolation and identification, as well as pharmacology, but little is known about its molecular genetics, limiting its conservation and utilization. This study aims to obtain and characterize the complete chloroplast genome of P. sarmentosum and provide valuable genomic information for its phylogeny and molecular breeding.
The fresh leaves of P. sarmentosum were collected from the Yulin Normal University Horticultural Germplasm Center in Yulin, Guangxi, China (110.185°E, 22.669°N). The voucher specimen (JL-YNU-001) was deposited in the Herbarium of Yulin Normal University (https://syy.ylu.cn/index.html, Yulin Zhu, [email protected]). A modified CTAB approach (Porebski et al. Citation1997) was used to extract the total genomic DNA of P. sarmentosum. Library construction and sequencing were conducted by Biozeron (Biozeron, Shanghai, China) using the Illumina HiSeq 4000 sequencing platform (Illumina, San Diego, CA) with 150 bp paired-end (PE) sequencing. The adapters and low-quality sequences of the obtained raw reads were trimmed and filtered by the Trimmomatic software (Bolger et al. Citation2014) with the options of ‘LEADING:2, TRAINLING:3, SLIDINGWINDOW:4:15, MINLEN:100.’ The de novo assembly and annotation of the P. sarmentosum chloroplast genome were achieved by the GetOrganelle toolkit (Jin et al. Citation2020) and CPGAVAS2 (Shi et al. Citation2019) with default parameters, respectively. The final chloroplast genome was submitted to GenBank with accession no. MZ958833.
The chloroplast genome, with a total length of 161,389 bp, has a typical quadripartite structure, containing a large single-copy (LSC) region of 88,979 bp, a small single-copy (SSC) region of 18,274 bp, and two copies of inverted-repeat regions (IRa and IRb) of 27,068 bp each. A total of 130 genes were annotated, comprising 85 protein-coding genes (PCGs), 8 ribosomal RNA (rRNA) genes, and 37 transfer RNA (tRNA) genes. 18 of these genes have double copies, and a total of 21 genes contain introns, including 8 tRNA genes and 13 PCGs, of which ycf3 and clpP contain two introns each, and the others contain only one intron.
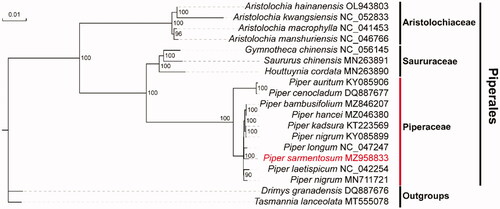
To confirm the phylogenetic position of P. sarmentosum, complete chloroplast genome sequences from the Piperaceae family and several closely related families from the order Piperales were used to reconstruct a phylogenetic tree. Multiple sequence alignments based on the single-copy orthologue genes from all chloroplast genomes were performed by MAFFT v7 (Katoh and Standley Citation2013) with default parameters, and the maximum-likelihood tree was built using IQ-TREE v1.6.10 (Nguyen et al. Citation2015) with the best-fit model TVM + F + G4 and 1000 bootstrap replicates (). Two species from the order Canellales, Drimys granadensis and Tasmannia lanceolata, were used as the outgroups. The phylogenetic tree shows that all Piperaceae species form a group, and P. sarmentosum in the current study is closely related to Piper longum. The complete chloroplast genome of P. sarmentosum could provide a genomic resource for further genetic and evolutionary studies on Piperales species.
Ethical approval
The data collection of plants was carried out with the permission of Yulin Normal University and complied with local (Yulin, Guangxin, China) legislation.
Authors’ contributions
Q. L. and R. C. were involved in the conception and design; X. G. and Y. Z. contributed the sample collection; X. G. and Z. R. performed the analysis and interpretation of the data; Q. L. and R. C. contributed the drafting of the paper; Q. L. and Y. Z. revised it critically for intellectual content. All authors were involved in the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ958833. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA818010, SRR18391781, and SAMN26814157, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):854–2120.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Mathew SP, Mohandas A, Nair G. 2004. Piper sarmentosum Roxb.–an addition to the flora of Andaman Islands. Curr Sci India. 87(2):141–142.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Sun X, Chen W, Dai W, Xin H, Rahmand K, Wang Y, Zhang J, Zhang S, Xu L, Han T. 2020. Piper sarmentosum Roxb.: a review on its botany, traditional uses, phytochemistry, and pharmacological activities. J Ethnopharmacol. 263:112897.