Abstract
The mitochondrial genome (mitogenome) of Ornithomya biloba (Dufour 1827) was first sequenced and annotated in this study as the first representative of the genus Ornithomya. The complete mitogenome is 18,654 bp in length and contains 37 genes (13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and control region). The phylogenetic analysis based on 13 PCGs in IQ-TREE supports the monophyly of Hippoboscidae, which was a sister group of Streblidae. Families Hippoboscidae and Streblidae formed the monophyletic Hippoboscoidea clade.
The family Hippoboscidae is characterized by the obviously dorsoventrally flattened body shape, head sunk into the thorax, their parasitism, and blood-sucking feeding habit (Soós and Hůrka 1986; Xue and Chao Citation1996). Hippoboscidae is a specific parasite family that infects birds and mammals (bats, cows, sheep, etc.) (Soós and Hůrka 1986). Some hippoboscid adults are vectors that could spread diseases (Xue and Chao Citation1996). Due to parasitic activity, they can spread worldwide with their hosts. There were 67 genera and 775 accepted species in the world (https://www.catalogueoflife.org/; query date: 2021-06-02). Hippoboscid flies are parasitic to birds and mammals and thus play an important role in their ecosystems. Here, we sequenced and annotated the mitochondrial genome data of Ornithomya biloba (Dufour Citation1827) in Hippoboscidae and roughly explored their phylogenetic relationship with some related groups.
The specimens of Ornithomya biloba (voucher number: LX2018-16) were collected in Longtan Waterfall, Wuling Mountain, Hebei Province, China (117.466003 E, 40.60182 N) by Jinlong Ren on 2 June 2018, and identified by Xin Li. The specimens were deposited in the Entomological Museum of China Agricultural University, Beijing, China (Liang Wang, [email protected]). The genomic DNA was extracted from the whole body (except wings) of the specimen using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), and then the DNA sample was stored at a −20 °C refrigerator. The mitochondrial genome was sequenced on the Illumina NovaSeq 6000 platform by Novogene Co., Ltd. (Cambridge, UK). Quality control and assembling were conducted in MitoZ software. A Python script circle_check.py in MitoZ software Github repository (https://github.com/linzhi2013/MitoZ) was used to confirm mitochondrial genome completeness (Meng et al. Citation2019). Annotation was executed in MITOS2 webserver (Donath et al. Citation2019) and corrected by hand following Cameron (Citation2014).
The complete mitochondrial genome (mitogenome) of Ornithomya biloba (GenBank accession number: MZ379837) is 18,654 bp, which contains 37 genes (13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and control region). The gene structure of O. biloba is similar to previous dipteran mitogenome studies (Zhou et al. Citation2017; Li et al. Citation2019). The nucleotide composition of O. biloba is 41.8% of A, 37.4% of T, 7.6% of G, and 13.2% of C, and A + T content is 79.2%. Six PCGs were started with ATG codon; atp8 was started with ATC; nad2, nad3, nad5, and nad6 were started with ATT; cox1 using TCG as start codon, and nad1 was initialed by TTG codon. Eight PCGs were terminated with TAA stop codon, while nad3 and cytb genes were stopped at TAG codon and cox1, cox2, nad5 were ended at single T. All tRNA genes were predicted and folded as cloverleaf structures.
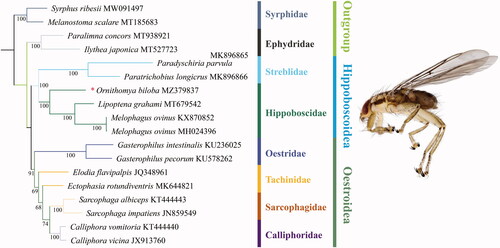
In our study, 13 PCGs of 18 species were used in phylogenetic analysis, the GenBank accession numbers are listed as follows: Syrphus ribesii MW091497 (Chen et al. Citation2021), Melanostoma scalare MT185683 (Liu et al. Citation2020), Paralimna concors MT938921 (Zhao et al. Citation2020), Ilythea japonica MT527723 (Wang et al. Citation2020), Paradyschiria parvula MK896865 (Trevisan et al. Citation2019), Paratrichobius longicrus MK896866 (Trevisan et al. Citation2019), Ornithomya biloba MZ379837 (present study), Lipoptena grahami MT679542 (Wang et al. Citation2021), Melophagus ovinus KX870852 (Liu et al. Citation2017), Melophagus ovinus MH024396 (Tang et al. Citation2018), Gasterophilus intestinalis KU236025 (Gao et al. Citation2016), Gasterophilus pecorum KU578262 (Zhang et al. Citation2016), Elodia flavipalpis JQ348961 (Zhao et al. Citation2013), Ectophasia rotundiventris MK644821 (Li et al. Citation2017), Sarcophaga albiceps KT444443 (Liao et al. Citation2016), Sarcophaga impatiens JN859549 (Nelson, Cameron, et al. Citation2012), Calliphora vomitoria KT444440 (Yan et al. Citation2014), Calliphora vicina JX913760 (Nelson, Lambkin, et al. Citation2012). All phylogenetic analyses were conducted in Phylosuite (Zhang et al. Citation2020), including sequences alignment in MAFFT (Katoh and Standley Citation2013), alignments trimming in trimAl (Capella-Gutierrez et al. Citation2009), substitution model selecting in ModelFinder (Kalyaanamoorthy et al. Citation2017), and maximum-likelihood phylogenetic tree rebuild in IQ-TREE (Minh et al. Citation2013; Nguyen et al. Citation2015). The topology and node support values are given in . IQ-TREE analysis revealed all the outgroups diverged from the rest. Newly sequenced Ornithomya biloba was sister to Lipoptena and Melophagus genera clade. All Hippoboscidae was monophyletic as a sister group of Streblidae. Oestrioidea and Hippoboscoidea were assigned to be sister groups in our dataset.
Authors’ contributions
XL and DY planned and designed the research. XL performed experiments, and XL and LW analyzed the data. XL wrote and DY revised the manuscript. All authors have approved the manuscript for publication and agreed to be accountable for all aspects of the work.
Ethics statement
The specimen collection protocol was approved by the Ethics Committee of China Agricultural University. The field collection permission was authorized by Wuling Mountain National Nature Reserve Management Committee and the collected insect samples are non-protected species.
Disclosure statement
All authors have read and approved the final manuscript. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The mitochondrial genome used in this study is available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number MZ379837. The associated BioProject, BioSample, and SRA numbers are PRJNA734480, SAMN19493608, and SRR14710877, respectively.
Additional information
Funding
References
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39(3):856–411.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Chen M, Peng K, Su C, Wang Y, Hao J. 2021. The complete mitochondrial genome of Syrphus ribesii (Diptera: Syrphoidea: Syrphidae). Mitochondrial DNA B Resour. 6(2):519–521.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Dufour L. 1827. Description et figure d'une nouvelle espèce d'ornithomyie. Ann Sci Nat. 10:243–248.
- Gao DZ, Liu GH, Song HQ, Wang GL, Wang CR, Zhu XQ. 2016. The complete mitochondrial genome of Gasterophilus intestinalis, the first representative of the family Gasterophilidae. Parasitol Res. 115(7):2573–2579.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li X, Ding SM, Hou P, Liu XY, Zhang CT, Yan D. 2017. Mitochondrial genome analysis of Ectophasia roundiventris (Diptera, Tachinidae). Mitochondrial DNA B Resour. 2(2):457–458.
- Li X, Wang L, Li Z, Hou P, Yang D. 2019. The mitochondrial genome of Formicosepsis sp. (Diptera: Cypselosomatidae). Mitochondrial DNA B Resour. 4(2):2140–2141.
- Liao H, Yang X, Li Z, Ding Y, Guo Y. 2016. The complete mitochondria genome of Parasarcophaga albiceps (Diptera: Sarcophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4696–4698.
- Liu ZQ, Kuermanali N, Li Z, Chen SJ, Wang YZ, Tao H, Chen CF. 2017. The complete mitochondrial genome of the parasitic sheep ked Melophagus ovinus (Diptera: Hippoboscidae). Mitochondrial DNA B Resour. 2(2):432–434.
- Liu D, Wang J, Song P, Li Y, Xie J, Yan J. 2020. Characterization of the complete mitochondrial genome of Melanostoma scalare (Diptera: Syrphidae). Mitochondrial DNA B Resour. 5(2):1753–1754.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nelson LA, Cameron SL, Yeates DK. 2012. The complete mitochondrial genome of the flesh fly, Sarcophaga impatiens Walker (Diptera: Sarcophagidae). Mitochondrial DNA. 23(1):42–43.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511(2):131–142.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Soós Á, Hůrka K. 1986. Family Hippoboscidae. In Soós Á, editor. Catalogue of Palaearctic Diptera. Vol. 11. Scathophagidae–Hypodermatidae. Budapest: Akadémiai Kiadó; p. 215–234.
- Tang JM, Li F, Cheng TY, Duan DY, Liu GH. 2018. Comparative analyses of the mitochondrial genome of the sheep ked Melophagus ovinus (Diptera: Hippoboscidae) from different geographical origins in China. Parasitol Res. 117(8):2677–2683.
- Trevisan B, Alcantara DMC, Machado DJ, Marques FPL, Lahr DJG. 2019. Genome skimming is a low-cost and robust strategy to assemble complete mitochondrial genomes from ethanol preserved specimens in biodiversity studies. PeerJ. 7:e7543.
- Wang M, Wang J, Guo Y, Zheng Q, Nouhoum D, Meng F. 2021. Complete mitochondrial genome of a potential vector louse fly, Lipoptena grahami (Diptera, Hippoboscidae). Mitochondrial DNA B Resour. 6(6):1752–1753.
- Wang L, Zeng W, Zhang J, Yang D. 2020. The mitochondrial genome of Ilythea japonica (Diptera: Ephydridae). Mitochondrial DNA B Resour. 5(3):2563–2564.
- Xue W, Chao C. 1996. China files. Vol. 2. Shenyang: Liaoning Science and Technology Press; p. 1366–2425.
- Yan J, Liao H, Zhu Z, Xie K, Guo Y, Cai J. 2014. The complete mitochondria genome of Calliphora vomitoria (Diptera: Calliphoridae). Mitochondrial DNA A. 1:1–379.
- Zhang D, Gao FL, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhang D, Yan L, Zhang M, Chu H, Cao J, Li K, Hu D, Pape T. 2016. Phylogenetic inference of calyptrates, with the first mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). Int J Biol Sci. 12(5):489–504.
- Zhao Z, Su TJ, Chesters D, Wang SD, Ho SYW, Zhu CD, Chen XL, Zhang CT. 2013. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of tachinid flies. PLOS One. 8(4):e61814.
- Zhao C, Wang J, Zhang B, Han J, Zhang J, Zhang D, Wang L. 2020. The mitochondrial genome of Paralimna (Paralimna) concors (Diptera: Ephydridae). Mitochondrial DNA B Resour. 5(3):3717–3718.
- Zhou Q, Ding S, Li X, Zhang T, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA B Resour. 2(2):461–462.