Abstract
Gymnogobius laevis (Steindachner, 1879) is a small, benthic freshwater fish. In a previous study, it was reported as a rare resident in Heilongjiang Province. In this study, the mitochondrial genome of G. laevis was sequenced. It is 16,519 base pairs (bp) in length and contains 13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA), and two non-coding control regions. The A + T content is 55.71% in the G. laevis mitochondrial genome. A phylogenetic tree shows that G. laevis is closely related to the family Gobiinae based on complete mitogenome sequences.
Gymnogobius laevis (Steindachner, 1879) is a small, benthic freshwater fish that belongs to the family Gobiinae of Perciformes. Gobiinae is one of the largest families in vertebrates (Nelson Citation2006). Most Gobiinae species like to live in warm waters, and several species live in freshwater rivers. G. laevis has been rarely found in Heilongjiang Province. In this study, G. laevis specimens were collected from the downstream of Tangwang River (E129°43′38.82″; N46°41′13.92″) in Heilongjiang Province, and its mitochondrial genome was sequenced and analyzed. This study of G. laevis has further enriched the fish resources that could provide basic information for other related studies.
All animal experiments were conducted in accordance with the guidelines and approval of the Animal Research and Ethics Committees of Harbin Sport University (approval number HTY20200711). The fins of G. laevis were collected for DNA extraction and the sample was stored in the herbarium of the Heilongjiang River Fisheries Research Institute (accession number: HRFRI-S-2021020; specimen ID: https://www.hrfri.ac.cn/; Li Peilun; [email protected]). DNA was collected according to the cetyltrimethylammonium bromide (CTAB) DNA extraction protocol (Zhai et al. Citation2021). The complementary DNA (cDNA) library was constructed and sequenced by using the Illumina NovaSeq at Jin Bei Biotech (Harbin, China). Approximately, 4.5 Gb of raw data were obtained, and the clean reads (29,720,032) were spliced using the SPAdes software (version 3.13) (Bankevich et al. Citation2012). The mitochondrial genome of G. laevis was annotated using MITOS (http://mitos.bioinf.uni-leipzig.de; Bernt et al. Citation2013). The mitochondrial sequence has been deposited in GenBank under the accession number MW049117.
The mitochondrial DNA sequence is 16,519 bp in length and contains 13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA), and two non-coding control regions. The A + T content is 55.71%. The 13 protein-coding genes are encoded on the light strand, except for ND6; these genes vary from 297 to 1554 bp in length. The genes ND1, ND2, COX2, ATP9, ATP6, COX3, ND3, ND4L, ND4, ND5, ND6, and CYTB begin with ATG as the start codon, while COX3 begins with GTG. The 22 tRNA genes are from 66 bp (tRNA-Phe) to 75 bp (tRNA-Lys) in length; they present a typical secondary structure. The two rRNA genes are 949 and 1637 bp. The 12S rRNA gene is located between tRNA-Phe and tRNA-Val, and the 16S rRNA gene is located between tRNA-Val and tRNA-Leu. The two non-coding control regions are 31 and 508 bp.
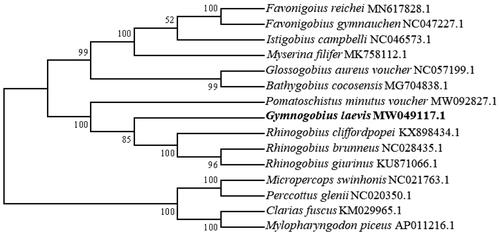
The mitochondrial DNA was submitted to multiple alignments by using Clustal W 2.0. The phylogenetic tree was constructed by using the maximum-likelihood method under the Tamura-Nei model, with 1000 bootstrap replicates in the MEGA 6.0 software based on the nucleotide sequences (Tamura et al. Citation2013). The mitochondrial genome sequences are used from 10 species in the Gobiinae family and three other families – Odontobutidae, including Micropercops swinhonis (Gunther, 1873) and Perccottus glenii (Dybowski, 1877); Siluriformes, including Clarias fuscus (Lacepede, 1803); and Cyprinidae, including Mylopharyngodon piceus (Richardson, 1846), which was used as an outgroup (Mascolo et al. Citation2018). G. laevis clusters with other members of the family Gobiinae and diverges from the other three families (). We hope that the present results help to clarify the taxonomic status of G. laevis: it is closely related to the family Gobiinae based on the mitochondrial genome.
Author contributions
Peng Lina: conceptualization, collection, software, and writing; Li Peilun: collection and preservation; Li Zheng and Li Hui: methodology, software, and data curation; Zhao Wenyan and Wang Hongkun: supervision, reviewing, and editing; Sun Lili: conceptualization, visualization, supervision, and editing.
Acknowledgements
The authors would like to thank the fisherman for the collecting of samples.
Disclosure statement
The authors have no potential conflict and are solely responsible for the content and writing of the manuscript.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (http://www.ncbi.nlm.nih.gov/) under accession no. MW049117.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA802364, SRR17659578, and SAMN 25538277, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):1–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018. The complete mitochondrial genome of the Pink dentex Dentex gibbosus (Perciformes: Sparidae). Mitochondrial DNA B Resour. 3(2):525–526.
- Nelson JS. 2006. Fishes of the world. New York: John Wiley and Sons, Inc.; p. 601.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Zhai H, Li Z, Mi S, Meng D, Yu H, Teng L, Liu Z. 2021. The complete mitochondrial genome of the Ferruginous Duck (Aythya nyroca) from Ningxia, China. Mitochondrial DNA B Resour. 6(2):546–547.