Abstract
The complete chloroplast genome sequence of Tainia acuminata Averyanov was assembled and the phylogenetic relationship of the species to other taxa in Subtrib. Bletlinae was inferred in this study. The length of the complete chloroplast sequence is 157,603 bp, and it contains a large single-copy (LSC) region of 86,336 bp, a small single-copy (SSC) region of 18,129 bp, and two inverted repeat (IRA and IRB) regions of 26,569 bp. A total of 134 genes were annotated including 89 protein-coding genes, 37 tRNA, and eight rRNA. Phylogenetic analysis showed that T. acuminata was closely related to T. cordifolia, and the genus was closely related to a clade consisting of Calanthe, Phaius, and Cephalantheropsis.
Tainia acuminata Averyanov Citation2013, a plant of Orchidaceae, is distributed in China and Vietnam (Averyanov Citation2013; Yuan et al. Citation2020). It has high ornamental value owing to its characteristic leaf shape and large beautiful flowers. T. acuminata has not been sequenced yet according to our knowledge, and the phylogenetic placement, species identification, genetic diversity, and conservation situation, etc. of the species remain unknown. The chloroplast sequences or genomes have been widely used in the studies of these areas (Daniell et al. Citation2016; Li et al. Citation2021). Here, we report the complete chloroplast genome of T. acuminata and its phylogenetic position in Subtrib. Bletlinae.
A sample of T. acuminata was collected from Jiuwanshan National Nature Reserve in Guangxi, China (25°11′26″ N, 108°47′31″ E), and the voucher specimen was deposited at the Herbarium of Guangxi Institute of Botany (http://www.gxib.cn/spIBK/, contact person: Chun-Rui Lin, Email: [email protected]) under the voucher number QY20190410027. The total DNA was extracted from silica-gel dried leaves by the CTAB method (Doyle and Doyle Citation1987). The genomic paired ends (PE150) sequencing was performed on NovaSeq 6000 (in Novogenecorp, Tianjin, China). Approximately, 1.6 Gb of clean data were gained after quality filtering using fastp (Chen et al. Citation2018). The complete chloroplast genome was de novo assembled using GetOrganelle (Jin et al. Citation2020), and annotated using PGA (Qu et al. Citation2019) and the online tool GeSeq (Tillich et al. Citation2017) with reference to the chloroplast genome of T. dunnii (NC_045862). The sequence was submitted to the GenBank (accession number: OL489753).
The complete chloroplast genome of T. acuminata has a length of 157,603 bp, consisting of a large single-copy (LSC 86,336 bp) region, a small single-copy (SSC 18,129 bp) region, and two inverted repeat (IRA and IRB 26,569 bp) regions. The overall GC content of the complete chloroplast genome is 37.3%. It contains 134 genes, including 89 protein-coding genes, 37 tRNA, and eight rRNA.
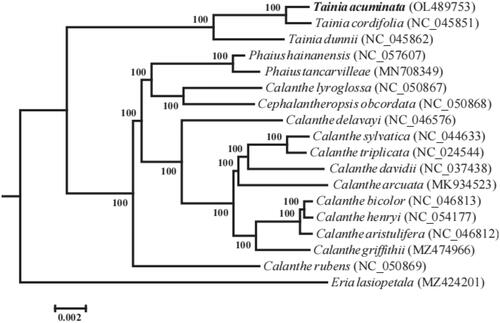
To investigate the phylogenetic position of T. acuminata, maximum-likelihood (ML) phylogenetic tree was reconstructed using RAxML (Stamatakis Citation2014) based on 17 chloroplast genomes using Eria lasiopetala as the outgroup. The phylogenetic tree indicated that T. acuminata was most closely related to T. cordifolia (BS = 100%) (), which was consistent with morphological observation (Averyanov Citation2013; Yuan et al. Citation2020). The genus was most closely related to a clade consisting of Calanthe, Phaius, and Cephalantheropsis.
Ethics statement
The sample collection was approved by Guangxi Jiuwanshan National Nature Reserve Administration.
Author contributions
Xia-lian Ou: analyzed data and wrote the manuscript. Ying Qin: collected materials and identified species. Li-guo Zhang: collected materials and analyzed data. Ting-guang Sun: identified and sent samples for sequencing. Xin-mei Qin: designed of the work and guided the analysis and modified the manuscript. All authors have read and agreed to publish the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OL489753. The associated BioProject, SRA, and BioSample numbers are PRJNA781090, SRR16964329, and SAMN23235068, respectively.
Additional information
Funding
References
- Averyanov LV. 2013. The orchids of Vietnam illustrated survey: Part 4. Subfamily Epidendroideae (tribes Arethuseae and Malaxideae). Turczaninowia. 16(1):884–163.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics. 34(17):i884–i890.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Li F, Xie X, Huang R, Tian EW, Li C, Chao Z. 2021. Chloroplast genome sequencing based on genome skimming for identification of Eriobotryae folium. BMC Biotechnol. 21(1):69.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15:50.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yuan Q, Tan F, Qin Y, Liu L. 2020. Tainia acuminata, a newly recorded species of Orchidaceae from China. J Trop Subtrop Bot. 28(3):245–247.