Abstract
Tylopilus plumbeoviolaceoides T.H. Li, B. Song & Y.H. Shen, 2002 is a species of basidiomycete in the family Boletaceae and is mainly found in Yunnan and Guangdong provinces in China. In this study, the mitochondrial genome of T. plumbeoviolaceoides was reported for the first time. The total length of the mitochondrial genome of T. plumbeoviolaceoides was 37,242 bp, with GC content of 23.0%. The mitochondrial genome of T. plumbeoviolaceoides contained 14 conserved protein-coding genes, 25 transfer RNA genes, and 2 ribosomal RNA genes. The phylogenetic tree indicated that T. plumbeoviolaceoides was closely related to Xerocomus impolitus and Butyriboletus roseoflavus. The complete mitochondrial genome of T. plumbeoviolaceoides will be useful for future research on basidiomycetes.
Tylopilus plumbeoviolaceoides T.H. Li, B. Song & Y.H. Shen, 2002 is a species of basidiomycete in the family Boletaceae and is mainly found in Yunnan and Guangdong provinces in China. A purple cap and a white stalk are distinguishing features of T. plumbeoviolaceoides (Falandysz et al. Citation2016; Rodríguez-Ramírez et al. Citation2020). Tylopilus plumbeoviolaceoides has a bitter taste and may cause diarrhea (Gelardi et al. Citation2015). The mitochondrial genome has a wide range of applications in the study of fungal species phylogeny (Zhang et al. Citation2017). However, the mitochondrial genome of T. plumbeoviolaceoides has not yet been reported. In this study, the mitochondrial genome of T. plumbeoviolaceoides was reported for the first time. We analyzed the general features of the mitochondrial genome of T. plumbeoviolaceoides and performed a phylogenetic analysis.
The specimen of T. plumbeoviolaceoides was sampled from Panlong District, Kunming City, Yunnan Province, China (24°23'N, 102°10'E). This research was conducted with the permission of the local government and the Kunming Institute of Botany, Chinese Academy of Sciences. The voucher samples and genomic DNA were stored at Qingdao University of Science and Technology (Chao Shi, [email protected]) under the specimen code TP0202109. The 500 bp genome sequencing libraries were constructed for de novo sequencing. These libraries were sequenced on the Illumina HiSeq platform (Illumina, San Diego, CA, USA) in Novogene (Beijing, China). The complete mitochondrial genome of T. plumbeoviolaceoides was assembled using NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017) and was annotated by MFannot that was used in previous studies (Zhang et al. Citation2017). The software of tRNAscanSE v1.21 (Schattner et al. Citation2005) was used to detect tRNA genes under the default settings, and RNAmmer (Lagesen et al. Citation2007) was used to validate rRNA genes under the default settings. Sequin was used to manually correct codons and gene boundaries.
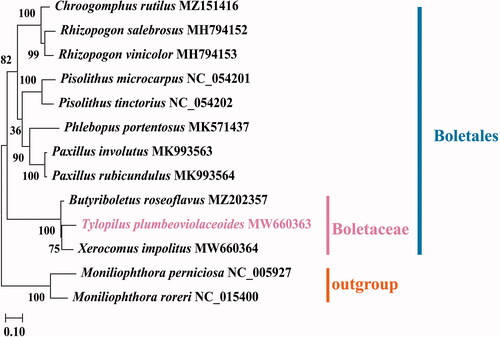
The complete mitochondrial genome of T. plumbeoviolaceoides was a typical circular molecule of 37,242 bp in length, with GC content of 23.0% (GenBank accession MW660363). The mitochondrial genome of T. plumbeoviolaceoides contained 20 putative protein-coding genes, including 14 conserved protein-coding genes (PCGs) and 6 open reading frames (ORFs) of unknown function. There were 25 transfer RNA (tRNA) genes in the mitochondrial genome of T. plumbeoviolaceoides, with 20 of which were unique. The 25 tRNA genes used 22 codons covered all 20 standard amino acids. There were two ribosomal RNA (rRNA) genes, small ribosomal RNA (rns) and large ribosomal RNA (rnl). The mitochondrial genome had a base composition of A (39.4%), C (11.4%), G (11.6%), and T (37.6%). The 14 conserved proteins were comprised of 7 NAD subunits dehydrogenases (nad1–6 and nad4L), 3 cytochrome oxidases (cox1–3), apocyto chrome b (cob), and 3 ATP synthases (atp6, apt8, and apt9). We constructed A phylogenetic tree based on mitochondrial protein-coding genes of 12 Boletales species from the NCBI database to reveal the phylogenetic relationships between T. plumbeoviolaceoides and other Boletales species. Moniliophthora perniciosa and Moniliophthora roreri were used as outgroup. To create sequence alignments for the construction of phylogenetic trees, MAFFT v725 (Katoh and Standley Citation2013) was applied to protein-coding genes. Then, the GTR-GAMMA (GTR + G) model was identified as the best fitting substitution model by applying the Bayesian Information Criterion (BIC) using Modeltest (Posada and Crandall Citation1998). Lastly, phylogenetic trees, with branch support based on 1000 bootstrap replicates, were inferred using the maximum likelihood (ML) method as implemented in MEGA-X software (Kumar et al. Citation2018). The phylogenetic tree indicated that T. plumbeoviolaceoides was closely related to Xerocomus impolitus and Butyriboletus roseoflavus than to other Boletales species (). The mitochondrial genome sequence of T. plumbeoviolaceoides will contribute to future research on basidiomycetes.
Author contributions
Wenbo Shi: Conceptualization, Data curation, Writing - Original Draft. Weicai Song: Methodology, Software. Yuan Peng: Data Curation. Shuo Wang: Formal analysis, Data Curation. Guiwen Yang: Investigation, Data Curation. Chao Shi: Data curation, Resources, Writing- Reviewing and Editing. All authors have read and approved the final manuscript.
Acknowledgements
We are thankful to Beijing-based Novogene for their NGS service that was instrumental to the execution of the project. We are grateful to Yongjie Lu for his guidance and assistance in the assembly of the mitochondrial genome of Tylopilus plumbeoviolaceoides in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession MW660363. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA786973, SRR17162356, and SAMN23765445, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: De Novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):999–9.
- Falandysz J, Saba M, Liu HG, Li T, Wang JP, Wiejak A, Zhang J, Wang YZ, Zhang D. 2016. Mercury in forest mushrooms and topsoil from the Yunnan Highlands and the subalpine region of the Minya Konka summit in the Eastern Tibetan plateau. Environ Sci Pollut Res Int. 23(23):23730–23741.
- Gelardi M, Vizzini A, Ercole E, Taneyama Y, Li TH, Zhang M, Yan WJ, Wang WJ. 2015. New collection, iconography and molecular evidence for Tylopilus Neofelleus (Boletaceae, Boletoideae) from Southwestern China and the taxonomic status of T. Plumbeoviolaceoides and T. Microsporus. Mycoscience. 56(4):373–386.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Rodríguez-Ramírez EC, Martínez-González CR, González-Ávila PA, Luna-Vega I. 2020. Tylopilus Hayatae, a new endemic bolete species in relict Mexican Beech forest. Phytotaxa. 441(1):35–46.
- Schattner P, Brooks AN, Lowe TM. 2005. The TRNAscan-SE, Snoscan and SnoGPS web servers for the detection of TRNAs and SnoRNAs. Nucleic Acids Res. 33(Web Server):W686–89.
- Zhang S, Wang XN, Zhang XL, Zhong Liu X, Zhang YJ. 2017. Complete mitochondrial genome of the endophytic fungus Pestalotiopsis Fici: features and evolution. Appl Microbiol Biotechnol. 101(4):1593–1604.
- Zhang YJ, Zhang HY, Liu XZ, Zhang S. 2017. Mitochondrial genome of the nematode endoparasitic fungus Hirsutella Vermicola reveals a high level of synteny in the family Ophiocordycipitaceae. Appl Microbiol Biotechnol. 101(8):3295–3304.