Abstract
Complete mitochondrial genomes of four species of Ethiopian speckled brush-furred rats Lophuromys (L. chrysopus, L. menageshae, L. melanonyx, and L. simensis) were assembled for the first time. We provide data concerning the sequencing, assembly, and annotation of the obtained mitogenomes; compare two widely used circular-genome annotation tools (MITOS and MitoZ), and discuss relevant points concerning relationships within both Ethiopian Lophuromys and the Muridae family.
Introduction
Ethiopian speckled brush-furred rats of the genus Lophuromys belong to the L. flavopunctatus species complex, which is widespread in Ethiopia and consists of nine species that are well-delimited both morphologically and genetically (Lavrenchenko et al. Citation2007; Bryja et al. Citation2019). The complex has emerged and flourished in the Ethiopian highlands, which are characterized by a patchwork of climatic conditions (Bryja et al. Citation2018). These circumstances have resulted in a process of adaptive radiation through so-called ‘reticulate’ evolution, which features multiple exchanges of genomic segments, primarily mitochondrial genomes, between species (Lavrenchenko et al. Citation2004; Komarova et al. Citation2021). As a consequence, among Ethiopian representatives of the genus, one can more or less clearly distinguish traces of at least four such introgression events, and some of them represent a complete replacement of the mitochondrial genome (Kostin et al. Citation2019). Despite an ever-increasing number of complete mitochondrial genomes, publicly available in the GenBank database, until the current study there was no mitochondrial genome data for none of Lophuromys genus representatives but only single cytb sequences (1140 bp). To fill this gap, we present here complete mitochondrial genomes of four out of the nine Ethiopian Lophuromys species [L. chrysopus Osgood, 1936; L. menageshae Lavrenchenko et al., 2007; L. melanonyx Petter, 1972 and L. simensis Osgood, 1936]. It is worth noting that in populations of the last two species, aside from the apparently species-specific mitotypes presented here, there is another one, presumably derived from L. menageshae [Kostin et al. (Citation2019); for more details, kindly refer to Komarova et al. (Citation2021)].
Materials and methods
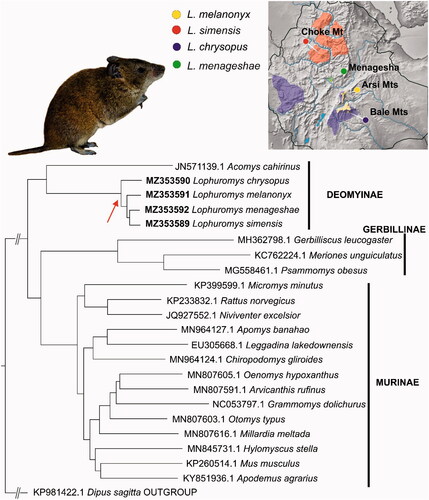
Muscle tissues were collected during the Joint Ethio-Russian Biological Expedition in 1998 (L. menageshae, Ethiopia, Menagesha forest, 8.95 N, 38.55 E), 2013 (L. chrysopus, Ethiopia, Bale Mts., Harenna forest, 6.645 N, 39.733 E), 2015 (L. melanonyx, Ethiopia, Arsi Mts., 7.825 N, 39.412 E,) and 2018 (L. simensis, Ethiopia, Choqe Mt., 10.7058 N, 37.8432 E) (see also , colored map). All experimental procedures were carried out in accordance with relevant guidelines in compliance with the International Union for Conservation of Nature (IUCN) policies regarding research involving species at risk of extinction (for details see Guidelines for appropriate uses of IUCN Red list data). All voucher specimens were deposited in the collection of the Zoological Museum of Lomonosov Moscow State University (https://zmmu.msu.ru, Vladimir S. Lebedev, [email protected]) under ID numbers S-165969 (L. menageshae), S-192730 (L. chrysopus), S-197618 (L. melanonyx) and S-202836 (L. simensis).
Figure 1. The maximum-likelihood phylogenetic tree constructed in IQTREE with 1000 bootstrap replications. Bootstrap supports of all nodes are >90. Red arrow shows the position of the Ethiopian Lophuromys on the phylogenetic tree. Colored map: schematic representation of geographic ranges of the four Lophuromys species. The circles show the voucher specimens’ collection sites.
DNA was extracted using a commercial kit (Jena Bioscience) according to the manufacturer’s instructions. Total genomic DNA was sonicated, and after adapter ligation, was sequenced on the Illumina HiSeq platform. Complete mitochondrial genomes were assembled by means of NOVOplasty 4.3.1 (Dierckxsens et al. Citation2017) with available sequences of the cytb gene serving as a starting seed. Because one of the aims of this study was the comparison of available annotation software, we tested two popular tools: MITOS (Bernt et al. Citation2013) and MitoZ (Meng et al. Citation2019). Finally, using complete mitochondrial sequences (available in GenBank) from 18 representatives of the Muridae family and the sequence of the northern three-toed jerboa (Dipus sagitta, Pallas, 1773) as an outgroup, we performed phylogenetic reconstruction by the maximum-likelihood approach in IQTREE (Nguyen et al. Citation2015) using 1000 bootstrap replications to assess robustness of the nodes.
Results and discussion
The complete mitogenomes of all four species were found to have a similar composition (L. simensis – 16,277 bp, L. chrysopus – 16,276 bp, L. melanonyx – 16,273 bp, and L. menageshae – 16,274 bp.) and contain 22 transfer RNA (tRNA) genes, two ribosomal-RNA genes (12S rRNA and 16S rRNA), 13 protein-coding genes (PCGs) and one non-coding control region. The overall nucleotide composition of the L. menageshae genome (in the other three species, it is virtually identical, with slight differences) is 33.7% of A, 26.6% of T, 12.2% of G, and 27.5% of C. The total length of the 13 PCGs is 11,310 bp. Initiation codons for all 13 PCGs can be described as ATN: ATA for nad1, nad2, and nad5; ATG for cox1, cox2, atp8, atp6, cox3, nad3 (L. chrysopus), nad4l, nad4, nad6, and cytb; and ATC for nad2 (L. simensis) and nad3 (L. chrysopus). Termination codons for all 13 PCGs are represented by two typical variants: either TAA or TAG. Both annotation tools (MITOS and MitoZ) revealed that the obtained mitogenomes are identical in structure (2 rRNA genes, 13 PCGs, and 22 tRNAs genes). Nonetheless, we found that MITOS has a weaker capacity for precise determination of gene length, especially at an end. For example, all 13 PCG sequences annotated by MITOS were shorter by 1–4 amino acid residues at the 3′ end. It is also worth mentioning that this MITOS drawback does not allow to deposit of obtained sequences in the GenBank database owing to the erroneous annotation. Until we identify the cause of errors in termination codon identification in MITOS, we cannot recommend this tool for annotation purposes and instead give our preference to MitoZ as more accurate. The mitogenomes annotated by means of MitoZ as well as the raw sequence reads were deposited in GenBank (see accession numbers below).
The resultant phylogenetic reconstruction () revealed the following pattern of relationships among the species: the basal position is occupied by L. chrysopus, and the next split divided L. melanonyx from the sister pair L. menageshae and L. simensis. In a comparison with a recently published Ethiopian Lophuromys study (Komarova et al. Citation2021) – based on a set of ddRadSeq SNPs and sequences of the cytb gene – our data turned out to be more similar to the topology of the ddRadSeq SNP tree (‘chrysopus’(‘melanonyx’(‘menageshae’ & ‘simensis’))) than to the tree based on a single mitochondrial gene (cytb, 1140 bp): (‘chrysopus’(‘simensis’(‘melanonyx’ & ‘menageshae’))). Taking into account quite recent diversification of Ethiopian Lophuromys [no later than ca. 1.4 million years ago; see Komarova et al. (Citation2021)], one can assume that the phylogenetic signal in the stand-alone mitochondrial gene (cytb) is not sufficient to resolve phylogenetic relationships between closely related species that have emerged from through rapid adaptive radiation. This observation points to the applicability of complete mitogenome sequences to the unraveling of shallow phylogenetic relationships below the genus level.
The other point of interest is that the obtained topology of the Muridae family is different from the generally accepted one [for example, see studies by Alhajeri et al. (Citation2015) and Aghová et al. (Citation2018) conducted on the basis of multilocus data including both nuclear and mitochondrial gene fragments]. Instead of expected sister relationships of subfamilies Deomyinae and Gerbillinae, our data are suggestive of a basal position of Deomyinae and proximity of Gerbillinae to Murinae (). It should be noted that the latter pattern is in agreement with a recently published paper (Song et al. Citation2021), where on the basis of mitochondrial sequences of 13 PCGs and two rRNAs, a similar topology was shown. Considering that the reconstruction of such deep diversification events by means of complete mitochondrial sequences may yield results that are erroneous or characterized by low bootstrap support, we have no choice but wait for the publication of nuclear genomic data, which are expected to resolve this kind of issues [for a similar example, see Mikula et al. (Citation2021)].
Despite the limited utility of the small number of mitogenome sequences, the gradual accumulation of a substantial set of sequences from non-model species opens up big opportunities for further research. This is especially true in the case of such groups as Lophuromys, where previously described putatively adaptive introgression events (Komarova et al. Citation2021) require more detailed and data-intensive investigation. Last but not least, it is important to note the narrowness of geographic ranges of Ethiopian Lophuromys species, especially for the obligate forest dweller L. menageshae (, colored map). In view of the accelerating pace of anthropogenic transformation, it is easy to predict that Lophuromys populations or even whole species [such as L. melanonyx, currently listed as vulnerable by the International Union for Conservation of Nature (Kennerley and Lavrenchenko Citation2016)] will face the threat of extinction.
Author contributions
L.A.L conceived the study and provided funding; D.S.K. and L.A.L collected samples in the field; N.S.M. performed sequencing; V.A.K and D.S.K. assembled genomes and analyzed the data; V.A.K and D.S.K drafted the manuscript; L.A.L. revised the manuscript critically for intellectual content. All authors contributed to the editing of the manuscript, gave final approval for publication, and agreed to be held accountable for the work performed therein.
Ethical approval
The study was conducted in strict accordance with guidelines on the implementation of the IUCN policy statement on research involving great risk of extinction with special references to Scientific Collecting of Threatened species as well as with the terms of research permits issued by Ethiopian authorities (Ethiopian Wildlife Conservation Authority; Oromia Forest and Wildlife Enterprise; Environment, Forest and Wildlife Protection and Development Authority of the Amhara National Regional State) and following national laws (Permission no. EWCA 31/336/05 of 20 March 2013; no. DhBBBO/PA-138/1326 of 05 February 2015; and no. ZN/R/R/95/0179 of 02 April 2018).
Acknowledgments
The authors thank Dr. Andrey Darkov (Joint Ethio-Russian Biological Expedition) for managing the expedition and Dr. Mesele Yihune (Addis Ababa University) for helping with obtaining the permits. The English language was corrected by shevchuk-editing.com.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of the NCBI (https://www.ncbi.nlm.nih.gov/) under the accession numbers MZ353589 – MZ353592. The associated BioProject, SRA and Bio-Sample numbers are PRJNA763532; SRR15904659, SRR15904661, SRR15904660 and SRR15904662; and SAMN21442456, SAMN21442454, SAMN21442455 and SAMN21442453, respectively.
Additional information
Funding
References
- Aghová T, Kimura Y, Bryja J, Dobigny G, Granjon L, Kergoat GJ. 2018. Fossils know it best: using a new set of fossil calibrations to improve the temporal phylogenetic framework of murid rodents (Rodentia: Muridae). Mol Phylogenet Evol. 128:1001–111.
- Alhajeri BH, Hunt OJ, Steppan SJ. 2015. Molecular systematics of gerbils and deomyines (Rodentia: Gerbillinae, Deomyinae) and a test of desert adaptation in the tympanic bulla. J Zoolog Syst Evol Res. 53(4):312–330.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bryja J, Kostin D, Meheretu Y, Šumbera R, Bryjová A, Kasso M, Mikula O, Lavrenchenko LA. 2018. Reticulate Pleistocene evolution of Ethiopian rodent genus along remarkable altitudinal gradient. Mol Phylogenet Evol. 118:75–87.
- Bryja J, Meheretu Y, Šumbera R, Lavrenchenko LA. 2019. Annotated checklist, taxonomy and distribution of rodents in Ethiopia. J Vertebr Biol. 68(3):117–213.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kennerley R, Lavrenchenko L. 2016. Lophuromys melanonyx. IUCN Red List Threat Species. 20162016: e.T12351A22408287. https://doi.org/https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T12351A22408287.en. Accessed on 15 August 2021.
- Komarova VA, Kostin DS, Bryja J, Mikula O, Bryjová A, Čížková D, Šumbera R, Meheretu Y, Lavrenchenko LA. 2021. Complex reticulate evolution of speckled brush‐furred rats (Lophuromys) in the Ethiopian Centre of Endemism. Mol Ecol. 30(10):2349–2365.
- Kostin DS, Kasso M, Komarova VA, Martynov AA, Gromov AR, Alexandrov DY, Bekele A, Zewdie C, Bryja J, Lavrenchenko LA. 2019. Taxonomic and genetic diversity of rodents from the Arsi Mountains (Ethiopia). Mammalia. 83(3):237–247.
- Lavrenchenko LA, Verheyen E, Potapov SG, Lebedev VS, Bulatova NS, Aniskin VM, Verheyen WN, Ryskov AP. 2004. Divergent and reticulate processes in evolution of Ethiopian Lophuromys flavopunctatus species complex: evidence from mitochondrial and nuclear DNA differentiation patterns. Biol J Linn Soc. 83(3):301–316.
- Lavrenchenko LA, Verheyen WN, Verheyen E, Hulselmans J, Leirs H. 2007. Morphometric and genetic study of Ethiopian Lophuromys flavopunctatus Thomas, 1888 species complex with description of three new 70-chromosomal species (Muridae, Rodentia). Bull Inst R Sci Nat Belg Biol. 77:77–117.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Mikula O, Nicolas V, Šumbera R, Konečný A, Denys C, Verheyen E, Bryjová A, Lemmon AR, Lemmon EM, Bryja J. 2021. Nuclear phylogenomics, but not mitogenomics, resolves the most successful Late Miocene radiation of African mammals (Rodentia: Muridae: Arvicanthini). Mol Phylogenet Evol. 157:107069.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Song SL, Yong H, Lim PE, Tan J. 2021. Mitochondrial genome of Rattus tiomanicus (Rodentia: Muridae) and molecular phylogeny of Murinae. Sains Malaysiana. 50(4):953–965.
