Abstract
The mitochondrial genome of Cylindromyia (Calocyptera) intermedia (Meigen, 1824) was sequenced and assembled through high-throughput sequencing techniques and reference-based assembly methods. The mitochondrial genome is 15,114 bp in total, consisting of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 noncoding control region. The nucleotide composition biases toward A and T; the overall A + T content are up to 76.9% of the entire mitogenome. The result of phylogenetic analysis suggested a close relationship between C. intermedia and subfamily Dexiinae.
Phasiinae is one of the four subfamilies of Tachinidae, however, there are only three mitochondrial genomes of this subfamily have been reported: Ectophasia rotundiventris (Loew, 1858), Gymnosoma dolycoridis (Dupuis, 1960) and Subclytia rotundiventris (Fallén, 1820) (Li et al. Citation2017; Pei et al. Citation2019). The genus Cylindromyia of tribe Cylindromyiini of subfamily Phasiinae includes 10 subgenera and 116 species worldwide, meanwhile 15 species are found in China (O’Hara et al. Citation2020). In this study, we sequenced the complete mitochondrial genome of Cylindromyia (Calocyptera) intermedia and analyzed its phylogenetic status, which provides useful genetic information for improving the taxonomic system and phylogenetics of Tachinidae.
The specimen (C. intermedia) was collected by sweeping collection from Huaxi District, Guiyang city, Guizhou Province, China (106.620159 N, 26.36755 E) on 7 June 2020 and was deposited at the Key Laboratory of Medical Insects of Guizhou Medical University (https://www.gmc.edu.cn/, Jiayu Liu, [email protected]) under the voucher number CI20200707. Total DNA was extracted from thoracic muscle tissues using Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, China). The sequencing library was established and further sequenced using the Illumina Hiseq PE150 platform, followed by additional bioinformatic analyses outlined below. The initial annotation of the mitogenome, including gene prediction and non-coding RNA, was conducted using MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Geneious Prime 2020.2.2 (Kearse et al. Citation2012) was used to compare the homologous gene annotations of the other two species of Phasiinae and then submitted data to GenBank database through NCBI.
The complete mitogenome of C. intermedia (GenBank accession number: OL539555) is 15,114 bp in length, with the following base composition: A (41%), T (35.9%), G (9%), and C (14.1%). It shows a conserved arrangement pattern, including 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes and one non-coding region. Four PCGs, two rRNA genes and eight tRNA genes are distributed in the light strand among the 38 sequence elements, while others are distributed in the heavy strand. The location and direction of all genes are conserved in respect to the other Phasiinae mitogenomes.
The 13 PCGs accounted for 73.9% of the complete mitogenome of C. intermedia (11,180 bp). PCGs utilize a variety of start codons including the standard ATN, except for the nonstandard CGA (COI) and TTG (ND1). The most frequent start codon was ATG, which was covered six PCGs (COII, ATP6, COIII, ND4, ND4L, CYTB). Nine PCGs (ND2, COI, COII, ATP8, ATP6, COIII, ND4L, ND6, CYTB) stop with TAA codon. ND3 and ND1 terminate with the codon TAG, while ND5 and ND4 have an incomplete stop codon T.
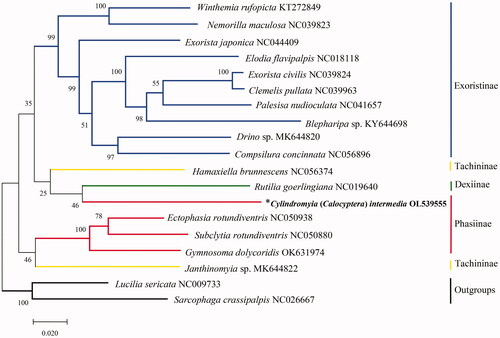
To investigate the phylogenetic status of C. intermedia in Phasiinae, a phylogenetic tree was reconstructed by the maximum-likelihood method using MEGAX 10.2.6 with bootstrap set to 1000 (Kumar et al. Citation2018) based on the combined 13 PCGs dataset for 17 tachinid species. Lucilia sericata (Meigen, 1826) from Calliphoridae and Sarcophaga crassipalpis Macquart, 1839 from Sarcophagidae were set as outgroups. The subfamily Phasiinae received a molecular phylogenetic treatment by Stireman et al. (Citation2019, Fig. 4), the clade tribe Cylindromyiini of Phasiinae sometimes joins the subfamily Dexiinae in individual gene-trees. Our phylogenetic analysis showed a close relationship between C. intermedia of Phasiinae and Rutilia goerlingiana of Dexiinae (), which was partially consistent with the previous work of Stireman et al. (Citation2019). The complete mitogenome of C. intermedia will contribute to the further studies on molecular bases for the classification and phylogeny of between Phasiinae and other three subfamilies within Tachinidae.
Ethical approval statement
The ethical approval (No. 1900074) was granted by the Animal Care Welfare Committee of Guizhou Medical University for the study.
Author contributions
Ming Yang and Jiayu Liu were involved in the conception and design; Rong Wang and Jiayu Liu analyzed and interpreted the data; Rong Wang drafted the article; Yan Zhi and Chuntian Zhang revised it critically for intellectual content. All authors approved the final version to be published and agreed to be accountable for all aspects of the work.
Disclosure statement
The authors report in this article without any conflict of interest. All authors agree to be accountable for all aspects of the work.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at under the accession no. OL539555. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA781126, SRR16980901, and SAMN23239677, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):1008–319.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li X, Ding SM, Hou P, Liu XY, Zhang CT, Yang D. 2017. Mitochondrial genome analysis of Ectophasia roundiventris (Diptera, Tachinidae)). Mitochondrial DNA B Resour. 2(2):457–458.
- O’Hara JE, Henderson SJ, Wood DM. 2020. Preliminary checklist of the Tachinidae (Diptera) of the world. Version 2.1. PDF document, 1039 pp.; [accessed 14 March 2022]. http://www.nadsdiptera.org/Tach/WorldTachs/Checklist/Tachchlist_ver2.1.pdf.
- Pei WY, Yan LP, Yang N, Zhang CT, Zheng CY, Yang J, Zhang D. 2019. First report of mitogenome of Subclytia rotundiventris (Diptera, Tachinidae) yielded by next-generation sequencing. Mitochondrial DNA B Resour. 4(2):2910–2911.
- Stireman JO, Cerretti P, O'Hara JE, Blaschke JD, Moulton JK. 2019. Molecular phylogeny and evolution of world Tachinidae (Diptera). Mol Phylogenet Evol. 139:106358.