Abstract
Utricularia tenuicaulis Miki 1935 is an aquatic carnivorous plant species found in East Asia including Korea and Japan. In this study, the chloroplast genome of U. tenuicaulis was successfully sequenced. The assembled genome (153,976 bp; GC ratio, 37.0%) contains four subregions, with the large single copy (LSC; 84,596 bp; 34.9%) and small single copy (SSC; 17,946 bp; 30.5%) regions separated by 25,718 bp of inverted repeat regions (42.7%), and includes 126 genes (81 protein-coding genes, 8 rRNAs, and 37 tRNAs). Phylogenetic analyses based on the whole-chloroplast genomes of 18 species, including 17 Lentibulariaceae species and one outgroup species, suggest a close relationship between U. tenuicaulis and Utricularia macrorhiza Leconte 1824. A comparison of genomic variation between U. tenuicaulis and U. macrorhiza confirmed the validity of the specific discrimination of U. tenuicaulis.
Aquatic bladderworts are carnivorous plants containing air-filled floating structures (Miki Citation1935; Rutishauser Citation2016). Utricularia tenuicaulis Miki Citation1935 was originally described as an aquatic bladderwort distinguished from U. japonica Mak. 1914 by its slenderer scape with a hollow core (Miki Citation1935; Shin et al. Citation2006). Taxonomically, U. japonica and U. tenuicaulis were merged into U. australis R. Br. 1810 as the sterile U. australis f. australis and the fertile U. australis f. tenuicaulis by Taylor (Citation1989), following the recommendation of Komiya and Shibata (Citation1980). In East Asia, the distribution of U. tenuicaulis closely overlaps those of U. japonica (= U. australis f. australis) and U. macrorhiza (Kadono Citation1994; Komiya Citation1997). Following detailed morphological investigation (Kadono Citation1994; Shin et al. Citation2006) and experimental confirmation of the origin of sterile U. australis through hybridization between U. tenuicaulis and U. macrorhiza (Kameyama et al. Citation2005), U. tenuicaulis was reconsidered as a distinct species. This taxonomic uncertainty prevented the full evaluation of the phylogenetic position of U. tenuicaulis in a recent phylogenetic study (Silva et al. Citation2018). Therefore, to investigate the taxonomic status of U. tenuicaulis, which has been misidentified as U. japonica in Korea (Na et al. Citation2008; Park, An, et al. Citation2020), we obtained its complete chloroplast genome sequences from a sample collected in Korea.
Total DNA (6.44 µg) was extracted from fresh leaves (600 mg) of U. tenuicaulis collected at the Saeteomal wetland at Gunpo-ro, Gyeonggi-do, Korea (37.339967°N, 126.936591°E) using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). A voucher specimen was deposited into the InfoBoss Cyber Herbarium (IN; http://herbarium.infoboss.co.kr/; voucher no., IBS-00023; Contact: Jongsun Park; [email protected]). Genome sequencing was conducted using a HiSeq4000 system at Macrogen Inc., Korea, and de novo assembly was performed using softwares; Velvet v1.2.10 (Zerbino and Birney Citation2008), GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) in the Genome Information System (GeIS) environment (http://geis.infoboss.co.kr/), which has been used in previous studies (Heo et al. Citation2020; Lee and Park Citation2021; Park, Kim, et al. Citation2021; Park, Min, et al. Citation2021). The Geneious Prime v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on the Pinguicula ehlersiae Speta & Fuchs chloroplast genome (NC_023463).
The U. tenuicaulis chloroplast genome (GenBank accession no. MN529625) is 153,976 bp in length, with a GC ratio of 37.0%, and has four subregions; the large single copy (LSC; 84,596 bp; 34.9%) and small single copy (SSC; 17,946 bp; 30.5%) regions separated by two inverted repeats (IRs; 25,718 bp; 42.7%), including 126 genes (81 protein-coding genes, 8 rRNAs, and 37 tRNAs) in the LSC and SSC regions and 17 genes (6 protein-coding genes, 4 rRNAs, and 7 tRNAs) duplicated in the IR regions. We determined four subregions by identifying junctions of two IR regions using the program, ‘BLAST 2 Sequences’ that supports BLAST searches to find the duplicated regions.
We identified 562 single-nucleotide polymorphisms (SNPs) and 302 indel regions (2,201 bp) with the comparison of U. macrorhiza (NC_025653). This indicates higher genomic variations in Utricularia species comparing to previous reports for other species; 220–520 SNPs and 125-144 indels in Castanopsis species (Park, Xi, et al. Citation2021), 7–27 SNPs and 19–49 indels in the Viburnum dilatatum species complex (Park, Xi, et al. Citation2020).
17 species in Lentibulariacea and Lippia origanoides Kunth. 1818 (Verbenaceae) as an outgroup were used for phylogenetic analysis. We used MEGAX (Kumar et al. Citation2018) to construct maximum-likelihood (ML) and neighbor-joining (NJ) trees and MrBayes v3.2.6 (Ronquist et al. Citation2012) to perform Bayesian inference (BI) after aligning the full chloroplast genomes using MAFFT v7.450 (Katoh and Standley Citation2013). We performed a heuristic search using nearest-neighbor interchange branch swapping, the Tamura–Nei model, and uniform rates among sites to construct ML and NJ phylogenetic trees, with default values for other options. To estimate node confidence, we performed bootstrap analyses with 1,000 and 10,000 pseudoreplicates for ML and NJ trees, respectively. For BI analysis, we used the general-time-reversible (GTR) model with gamma rates as the molecular model and a Markov chain Monte Carlo algorithm implemented for 1,100,000 generations. To build the BI consensus tree, we sampled trees every 200 generations after removing 100,000 generations as burn-in. All phylogenetic trees inferred from the ML, NJ, and BI methods showed the same topology, with three genera of Lentibulariaceae grouped with strong support (). Our phylogenetic analysis indicated that U. tenuicaulis is closely related to but distinguished from U. macrorhiza as the phylogenetic distance between two species is similar or larger than those among Genlisea species ().
Figure 1. Phylogenetic tree inferred from 18 chloroplast genomes representing 17 Lentibulariaceae species and one outgroup species. The maximum likelihood (ML) tree is presented with bootstrap support values and posterior probabilities estimated using the ML search, neighbor-joining, and Bayesian inference methods.
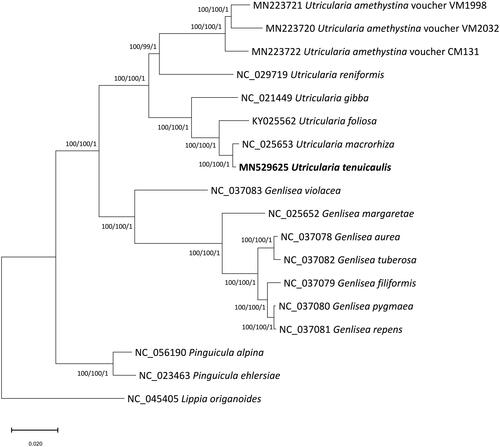
These results suggest that U. tenuicaulis is an independent taxon, genetically distinguished from U. macrorhiza. Further sequencing analysis including a wider range of taxa is necessary to clarify the phylogenetic relationships of Utricularian species in greater detail.
Ethical approval
Authors declare that there is no ethical or legal violation in obtaining the study materials and preforming research. The species used in this study is not listed in the IUCN Red List and plant materials were collected in the location that was not designated as a protective area in Korea. Authors confirmed that the plant materials for this study were not subjected to be approved from Institutional Review Board (IRB) in the Catholic University of Korea.
Author contributions
JP and STK conceptualized the project and designed the experiment. JP, YK generated sequencing data. JP, XH and STK analyzed the data. JP and STK wrote the manuscript with input from all other authors. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We thank Ms. Kumsoon Lee for her advice on the sampling location in Korea.
Disclosure statement
Jongsun Park and Xi Hong are employees of InfoBoss Inc. No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MN529625. The associated BioProject, Sequenced Read Archive, and Bio-Sample numbers are PRJNA764600, SAMN21509661, and SRR15970505 respectively.
Additional information
Funding
References
- Heo K-I, Park J, Xi H, Min J. 2020. The complete chloroplast genome of Agrimonia pilosa Ledeb. isolated in Korea (Rosaceae): investigation of intraspecific variations on its chloroplast genomes. Mitochondrial DNA B Resour. 5(3):2264–2266.
- Kadono Y. 1994. Aquatic plants of Japan. Tokyo, Japan: Bun-ichi Sogo Shuppan.
- Kameyama Y, Toyama M, Ohara M. 2005. Hybrid origins and F1 dominance in the free-floating, sterile bladderwort, Utricularia australis f. australis (Lentibulariaceae)). Am J Bot. 92(3):469–476.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Komiya S. 1997. Carnivorous plants in Hokkaido, northern Japan. Bullet Nippon Dental Univ Gen Educ. 26:153–188.
- Komiya S, Shibata C. 1980. Distribution of the Lentibulariaceae in Japan. Bullet Nippon Dental Univ Gen Educ. 9:163–212.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetlics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee B, Park J. 2021. The complete chloroplast genome of Zoysia japonica Steud. isolated in Korea (Poaceae): investigation of potential molecular markers on Z. japonica chloroplast genomes. Plant Biotechnol Rep. 15(5):707–709.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v1 [q-bio.GN].
- Miki S. 1935. New water plants in Asia Orientalis III. Shokubutsugaku Zasshi. 49(588):847–852.
- Na ST, Choi H-K, Kim YD, Shin H. 2008. Taxonomic identities and distribution of Utricularia japonica and U. tenuicaulis in Korea. Korean J Pl Taxon. 38(2):111–120.
- Park J, An J-H, Kim Y, Kim D, Yang B-G, Kim T. 2020. Database of National Species List of Korea: the taxonomical systematics platform for managing scientific names of Korean native species. J Species Res. 9(3):233–246.
- Park J, Kim Y, Kwon W, Xi H, Park C-H. 2021. The complete chloroplast genome sequence of new species candidate of Plantago depressa Willd. in Korea (Plantaginaceae). Mitochondrial DNA B Resour. 6(7):1961–1963.
- Park J, Min J, Kim Y, Chung Y. 2021. The comparative analyses of six complete chloroplast genomes of morphologically diverse Chenopodium album L.(Amaranthaceae) collected in Korea. Int J Genomics. 2021:6643444.
- Park J, Xi H, Oh S-h. 2020. Comparative chloroplast genomics and phylogenetic analysis of the Viburnum dilatatum complex (Adoxaceae) in Korea. Korean J Pl Taxon. 50(1):8–16.
- Park J, Xi H, Son J, Shin HT, Kang H, Park S. 2021. The complete chloroplast genome of Castanopsis sieboldii (Makino) Hatus (Fagaceae). Mitochondrial DNA B Resour. 6(9):2743–2745.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Rutishauser R. 2016. Evolution of unusual morphologies in Lentibulariaceae (bladderworts and allies) and Podostemaceae (river-weeds): a pictorial report at the interface of developmental biology and morphological diversification. Ann Bot. 117(5):811–832.
- Shin H, Kadono Y, Choi H-K. 2006. Taxonomic notes on the Dr. Miki's specimens collected from Korea. Korean J Pl Taxon. 36(1):41–52.
- Silva SR, Gibson R, Adamec L, Domínguez Y, Miranda VF. 2018. Molecular phylogeny of bladderworts: a wide approach of Utricularia (Lentibulariaceae) species relationships based on six plastidial and nuclear DNA sequences. Mol Phylogenet Evol. 118:244–264.
- Taylor P. 1989. The genus Utricularia. A taxonomic monograph. London. HMSO.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(S14):S2.
